1.
Introduction
Due to its broad relevance and propensity to incorporate many repercussions of actual concerns, the idea of fractional calculus (FC) has garnered considerable prominence in previous decades. Classical calculus has remained a small segment of FC, despite the fact that it can demonstrate numerous critical challenges and assist us in forecasting the behaviour of intricate occurrences in impulsive integro-differential equations [1], neural networking [2], thermal energy [3], non-Newtonian fluids [4] and heat flux [5]. Despite the fact that innovators offer numerous novel concepts, several aspects should always be deduced in order to guarantee all categories of phenomenon, that will be accomplished by conquering the restrictions posed by mathematicians and scientists. This is highly pertinent when investigating of MHD electro-osmotically flow [6], epidemics [7,8,9], stability and instability of special functions [10,11,12,13], inequalities [14,15,16] as well as other disciplines. When it tends to arrive to the exploration of repercussions that assist in resolving major difficulties (such as the current global challenges), there is always room for improvements, innovation, creativeness, and extensions in analysis, and so many investigators have inferred provoking outcomes with the assistance of FC, and by incorporating efficacious methodologies with the assistance of underlying FC findings [17,18,19].
Numerical models investigation and analysis of corresponding features are often a high priority in mathematical modeling when the relevant techniques are implemented. This is certainly pertinent in epidemic research, bifurcation, thermodynamics, electrostatistics modeling, fluid flow, plasma physics, and other fields. Several approaches, including N-solitons [20,21,22,23], solitary waves [24,25,26,27], Tan-Cot function method [28], Adomian decomposition method [29], homotopy perturbation method [30], q-homotopy analysis method [31], variation iteration method [32], collocation-shooting method [33], G/G′ expansion method [34], improved tan(ϕ(τ)/2)-expansion method [35], Lie symmetry analysis method [36], wavelet method [37] have been employed and refined by researchers to achieve the analytic, semi-analytic, and numerical solution of nonlinear PDEs. The Adomian decomposition method [29,38] is one of them, and it offers an efficient approach for exact-analytical solutions across a broad and comprehensive domain of specific aspects that simulate real-world issues. This strategy transforms a basic, incredibly straightforward problem into the complicated problem under investigation, and when combined with Adomian components, it offers a tremendous mathematical instrument. Numerous aspects of the Adomian decomposition method have also received considerable focus recently.
In this analysis, we examine a nonlinear framework that explains powerful interactions between interior disturbances in the water. The Korteweg de-Vries (KdV) equations are frequently utilized to illustrate acoustic wave behaviour and its physical relevance. Due to the immense amplitude of lengthy longitudinal waves and prolonged rotating impacts, we employ a modified KdV equation. We now evaluate the following models using the isopycnic surface W(x,t), which dipicts the KdV and modified KdV equations [39], respectively:
where a1 and a2 signifies the quadratic and cubic non-linear coefficients, respectively. Also, the coefficient of small-scale dispersion is denoted by b1. Here, a1 and a1 presents the proportional factors associating in the aforsaid equation, and is due to the nonlinear hydrodynamic system, and it appears classically, see[40,41].
Furthermore, we compute the exact-analytical solution of nonlinear dispersive equations K(n,n):
Equation (1.2) is the evolutionary model for compactons. Compactons are characterized as solitons with bounded wave lengths or solitons without exponential tails in solitary wave theory (Rosenau and Hyma, 1993). Compactons are formed by the intricate coupling of nonlinear convection ∂∂x(Wn) and nonlinear dispersion ∂3∂x3(Wn) in (1.2).
Amidst Gorge Adomian's massive boost in 1980, the Adomian decomposition method introduced a well-noted terminology. It has been intensively implemented for a diverse set of nonlinear PDEs, for instance, the Korteweg-De Vries model [42], Fisher's model [43], Zakharov–Kuznetsov equation [44] and so on. The ADM was determined to be significantly related to a variety of integral transforms, including Laplace, Swai, Mohand, Aboodh, Elzaki, and others. Humanity is continuously striving to improve performance and minimizing the method's intricacy through invention, modernity, and experimentation. In connection with this, Jafari [45] propounded a well-known integral transform which is known to generalized integral transform. The dominant feature of this transformation is that it has the ability to recapture several existing transformations, see Remark 1.
Motivated by the above propensity, we aim to establish a semi-analytical approach by mingling the Jafari transform with the Adomian decomposition method, namely the Jafari decompostion method (JDM). With the assistance of fractional derivative operators, we constructed the approximate-analytical solutions for KdV, MKdV, K(2,2) and K(3,3). The suggested methodology helps us increase flexibility in determining the initial conditions, and its novelty is that it has a straightforward solution technique. This approach is straightforward and encompasses all of JDM accomplishments, as well as encouraging several scholars to investigate a broad spectrum of applications and processes. The tool to overcome computational complexity without any constraints, perturbations, or transformations from nonlinear to linear, or partial to ordinary differential equations, is the distinctive characteristic of the proposed approach. Furthermore, it is connected to factors that are extremely useful in bringing the findings to a favourable conclusion. It also is coupled to well-posed transformation, which tries to diminish the technique's intricacy while increasing its application and dependability.
2.
Preliminaries
In this section, we evoke some essential concepts, notions, and definitions concerning fractional derivative operators depending on power and Mittag-Leffer as a kernel, along with the detailed consequences of the Jafari transform.
Definition 2.1. ([17]) The Caputo fractional derivative (CFD) is described as follows:
Definition 2.2. ([18]) The Atangana-Baleanu fractional derivative operator in the Caputo form (ABC) is stated as follows:
where W∈H1(a1,a2)(Sobolevspace),a1<a2,λ∈[0,1] and A(λ) signifies a normalization function as A(λ)=A(0)=A(1)=1.
Definition 2.3. ([18]) The fractional integral of the ABC-operator is described as follows:
Definition 2.4. ([45]) Consider an integrable mapping W(t) defined on a set P, then
Definition 2.5. ([45]) Suppose the mappings ϕ(s),ψ(s):R+↦R+ such that φ(s)≠0∀s∈R+. The Jafari transform of the mapping W(t) presented by Q(s) is described as
Theorem 2.6. ([45]) (Convolution property). For Jafari transform, the subsequent holds true:
Definition 2.7. The Jafari transform of the CFD operator is stated as follows:
Remark 1. Definition 2.7 leads to the following conclusions:
1) Taking ϕ(s)=1 and ψ(s)=s, then we acquire the Laplace transform [46].
2) Taking ϕ(s)1s and ψ(s)=1s, then we acquire the α-Laplace transform [47].
3) Taking ϕ(s)=1s and ψ(s)=1s, then we acquire the Sumudu transform [48].
4) Taking ϕ(s)=1s and ψ(s)=1, then we acquire the Aboodh transform [49].
5) Taking ϕ(s)=s and ψ(s)=s2, then we acquire the Pourreza transform [50,51].
6) Taking ϕ(s)=s and ψ(s)=1s, then we acquire the Elzaki transform [52].
7) Taking ϕ(s)=u2 and ψ(s)=su2, then we acquire the Natural transform [53].
8) Taking ϕ(s)=s2 and ψ(s)=s, then we acquire the Mohand transform [54].
9) Taking ϕ(s)=1s2 and ψ(s)=1s, then we acquire the Swai transform [55].
10) Taking ϕ(s)=1 and ψ(s)=1s, then we get the Kamal transform [56].
11) Taking ϕ(s)=sα and ψ(s)=1s, then we acquire the G−transform [57,58].
Definition 2.8. ([59]) The Jafari transform of the ABC fractional derivative operator is described as:
Remark 2. Definition 2.8 leads to the following conclusions:
1) Taking ϕ(s)=1 and ψ(s)=s, then we acquire the Laplace transform of ABC fractional derivative operator [60,61].
2) Taking ϕ(s)=s and ψ(s)=1s, then we acquire the Elzaki transform of ABC fractional derivative operator [62].
3) Taking ϕ(s)=ψ(s)=1s, then we get the Sumudu transform of ABC fractional derivative operator [63].
4) Taking ϕ(s)=1 and ψ(s)=s/u2, then we get the Shehu transform of ABC fractional derivative operator [63].
Definition 2.9. ([64]) The Mittag-Leffler function for single parameter is described as
3.
Description of the Jafari decomposition method involving singular and nonsingular kernels
Consider the generic fractional form of PDE:
with ICs
where Dλt=∂λW(x,t)∂tλ symbolizes the Caputo and ABC fractional derivative of order λ∈(0,1] while L and N denotes the linear and nonlinear factors, respectively. Also, F(x,t) represents the source term.
Taking into account the Jafari transform to (3.1), and we acquire
Firstly, applying the differentiation rule of Jafari transform with respect to CFD, then we apply the ABC fractional derivativ operator as follows:
and
The inverse Jafari transform of (3.3) and (3.4) yields
and
The generalized decomposition method solution W(x,t) is represented by the following infinite series
Thus, the nonlinear term N(x,t) can be evaluated by the Adomian decomposition method prescribed as
where
Inserting (3.7) and (3.8) into (3.5) and (3.6), respectively, we have
and
Consequently, the recursive technique for (3.9) and (3.10) are established as:
4.
Physical interpretation of Jafari decomposition method
In what follows, we present the various kinds of partial differential equations with the CFD and AB-frctional derivative operators, respectively.
4.1. Fractional KdV equation
Example 4.1. Assume that the time-fractional KdV equation
with IC:
Proof. Foremost, we provide the solution of (4.1) in two general cases.
Case Ⅰ. Firstly, we apply the Caputo fractional derivative operator coupled with the Jafari transform and Adomian decomposition method. Applying the Jafari transform to (4.1).
Taking into consideration the IC given in (4.2), we have
Employing the inverse Jafari transform, we obtain
Thanks to the JDM, we find
Here, we surmise that the unknown function W(x,t) can be written by an infinite series of the form
Also, the non-linearity F(W) can be decomposed by an infinite series of polynomials represented by
where Wℓ(x,t) will be evaluated recurrently, and Aℓ is the so-called polynomial of W0,W1,...,Wℓ established by [65].
The first few Adomian polynomials are presented as follows:
For ℓ=0,1,2,3,...
The approximate solution for Example 4.1 is expressed as:
Case Ⅱ. Here, we surmise ABC fractional derivative operator coupled with the Jafari transform and Adomian decomposition method. Applying the Jafari transform for Example 4.1.
Taking into consideration the IC given in (4.2), we have
Employing the inverse Jafari transform, we obtain
Thanks to the JDM, we find
Here, we surmise that the unknown function W(x,t) can be written by an infinite series of the form
Also, the non-linearity F1(W) can be decomposed by an infinite series of polynomials represented by
where Wℓ(x,t) will be evaluated recurrently, and Aℓ is the so-called polynomial of W0,W1,...,Wℓ defined in (4.5). Then, we have
For ℓ=0,1,2,3,...
The approximate solution for Example 4.1 is expressed as:
For λ=1, we obtained the exact solution of Example 4.1 as
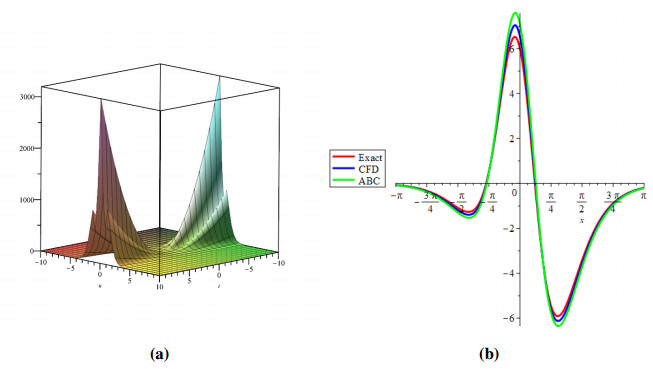
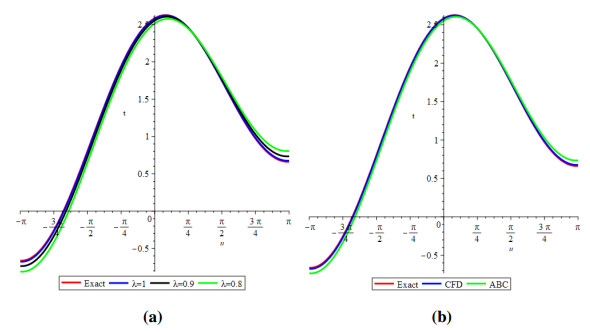
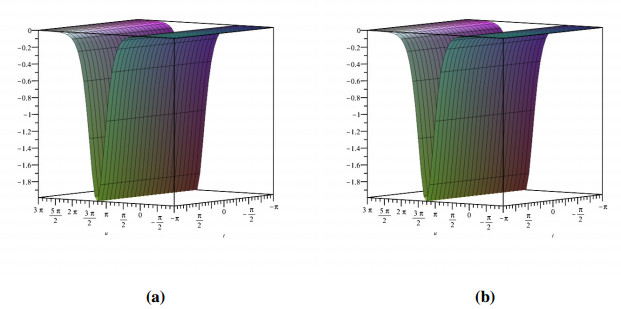
Figure 1 shows the evolutionary outcomes for the explicit and approximate solutions of Example 4.1 for the particular instance λ=1. The result generated by the proposed technique is remarkably similar to the exact solution, as shown in Figure 1.
Then, by considering only the first few elements of the linear equations features are integrated, we can deduce that we have accomplished a reasonable estimation with the numerical solutions of the problem. It is obvious that by adding additional components to the decomposition series (4.6) and (4.9), the cumulative error can be diminished.
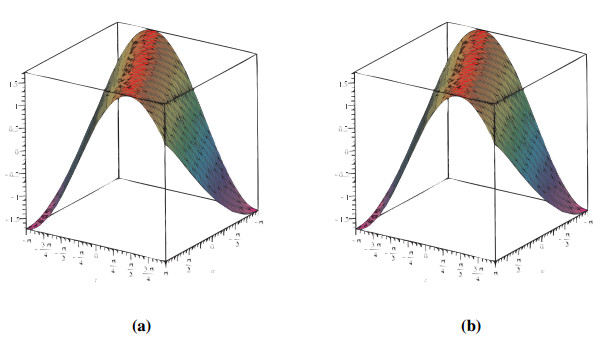
Analogously, we demonstrate the two-dimensional view of the change in fractional values of the order. We depict the response in Figure 2. It is remarkable that pairwise collisions of particle-like phenomena (including solitary waves and breathers) are fundamental mechanisms in the production of condensed soliton gas dynamics. Deep water waves, shallow groundwater waves, internally waves in a segmented sea, and fibre optics are all manifestations of these waves.
Remark 3. It is remarkable that equivalent version of the KdV equation is presented as
with IC:
has the solitary wave solution, when λ=1, then
4.2. Fractional modified KdV equation
Example 4.2. Assume that the time-fractional modified KdV equation
with IC:
Proof. Foremost, we provide the solution of (4.10) in two general cases.
Case Ⅰ. Firstly, we apply the Caputo fractional derivative operator coupled with the Jafari transform and Adomian decomposition method.
Applying the Jafari transform to (4.10).
Taking into consideration the IC given in (4.11), we have
Employing the inverse Jafari transform, we obtain
Thanks to the JDM, we find
Here, we surmise that the unknown function W(x,t) can be written by an infinite series of the form
Also, the non-linearity F(W) can be decomposed by an infinite series of polynomials represented by
where Wℓ(x,t) will be evaluated recurrently, and Bℓ is the so-called polynomial of W0,W1,...,Wℓ established by [65].
The first few Adomian polynomials are presented as follows:
For ℓ=0,1,2,3,...
The approximate solution for Example 4.2 is expressed as:
Case Ⅱ. Here, we surmise ABC fractional derivative operator coupled with the Jafari transform and Adomian decomposition method.
Applying the Jafari transform on (4.10).
Taking into consideration the IC given in (4.11), we have
Employing the inverse Jafari transform, we obtain
Thanks to the JDM, we find
Here, we surmise that the unknown function W(x,t) can be written by an infinite series of the form
Also, the non-linearity F(W), can be decomposed by an infinite series of polynomials represented by
where Wℓ(x,t) will be evaluated recurrently, and Bℓ is the so-called polynomial of W0,W1,...,Wℓ defined in (4.14).
For ℓ=0,1,2,3,...
The approximate solution for Example 4.2 is expressed as:
For λ=1, we obtained the exact solution of Example 4.2 as
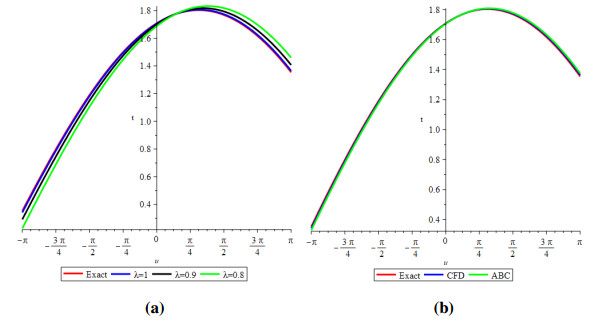
Figure 3 shows the evolutionary outcomes for the explicit and approximate solutions of Example 4.2 for the particular instance λ=1. The result generated by the proposed technique is remarkably similar to the exact solution, as shown in Figure 3.
Then, by considering only the first few elements of the nonlinear equations features are integrated, we can deduce that we have accomplished a reasonable estimation with the numerical solutions of the problem. It is obvious that by adding additional components to the decomposition series (4.15) and (4.18), the cumulative error can be diminished.
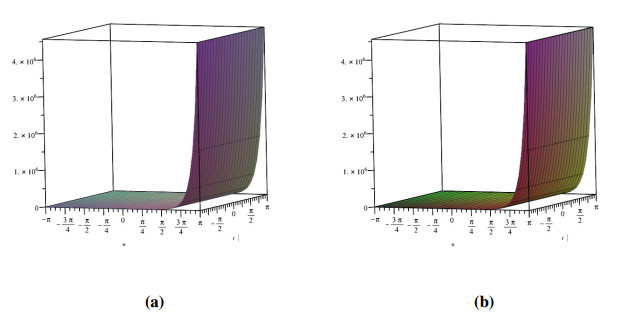
Analogously, we demonstrate the two-dimensional view of the change in fractional values of the order. Figure 4 depicts the response for exact CFD and AB fractional derivative operators. It is remarkable that pairwise collisions of particle-like phenomena (including solitary waves and breathers) are fundamental mechanisms in the production of condensed soliton gas dynamics. Deep water waves, shallow groundwater waves, internally waves in a segmented sea, and fibre optics are all manifestations of these waves.
4.3. Fractional K(2,2) equation
Example 4.3. Assume that the time-fractional K(2,2) equation
with IC:
Proof. Foremost, we provide the solution of (4.19) in two general cases.
Case Ⅰ. Firstly, we apply the Caputo fractional derivative operator coupled with the Jafari transform and Adomian decomposition method.
Applying the Jafari transform on (4.19).
Taking into consideration the IC given in (4.20), we have
Employing the inverse Jafari transform, we obtain
Thanks to the JDM, we find
Here, we surmise that the unknown function W(x,t) can be written by an infinite series of the form
Also, the non-linearities F1(W) and F2(W) can be decomposed by an infinite series of polynomials represented by
where Wℓ(x,t) will be evaluated recurrently, and Dℓ and Eℓ are the so-called polynomial of W0,W1,...,Wℓ established by [65].
The first few Adomian polynomials are presented as follows:
For ℓ=0,1,2,3,...
The approximate solution for Example 4.3 is expressed as:
Case Ⅱ. Here, we surmise ABC fractional derivative operator coupled with the Jafari transform and Adomian decomposition method. Applying the Jafari transform for Example 4.19.
Taking into consideration the IC given in (4.20), we have
Employing the inverse Jafari transform, we obtain
Thanks to the JDM, we find
Here, we surmise that the unknown function W(x,t) can be written by an infinite series of the form
Also, the non-linearity Fȷ(W),ȷ=1,2 can be decomposed by an infinite series of polynomials represented by
where Wℓ(x,t) will be evaluated recurrently, and Dℓ and Eℓ are the so-called polynomial of W0,W1,...,Wℓ established defined in (4.23). Then, we have
For ℓ=0,1,2,3,...
The approximate solution for Example 4.3 is expressed as:
For λ=1, we obtained the closed form solution of Example 4.3 as
Figure 5 shows the evolutionary outcomes for the explicit and approximate solutions of Example 4.3 for the particular instance λ=1. The result generated by the proposed technique is remarkably similar to the exact solution, as shown in Figure 5.
Then, by considering only the first few elements of the nonlinear equations features are integrated, we can deduce that we have accomplished a reasonable estimation with the numerical solutions of the problem. It is obvious that by adding additional components to the decomposition series (4.24) and (4.27), the cumulative error can be diminished.
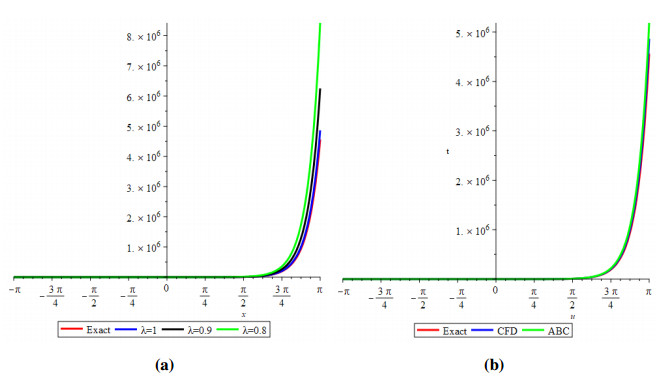
Analogously, we demonstrate the two-dimensional view of the change in fractional values of the order. Figure 6 depicts the response for exact CFD and AB fractional derivative operators.
For a variation of the K(2,2) equation, constrained traveling-wave solutions are achieved. We acquire hump-shaped and valley-shaped solitary-wave solutions, as well as some periodic solutions, for the focusing branch. It is worth noting that optimal focusing provides the aggregate of the frequency and amplitude of the originating waves in the engaging phase, as illustrated in reference [66].
4.4. Fractional K(3,3) equation
Example 4.4. Assume that the time-fractional K(3,3) equation
with IC:
Proof. Foremost, we provide the solution of (4.28) in two general cases.
Case Ⅰ. Firstly, we apply the Caputo fractional derivative operator coupled with the Jafari transform and Adomian decomposition method.
Applying the Jafari transform on (4.28).
Taking into consideration the IC given in (4.29), we have
Employing the inverse Jafari transform, we obtain
Thanks to the JDM, we find
Here, we surmise that the unknown function W(x,t) can be written by an infinite series of the form
Also, the non-linearities F1(W) and F2(W) can be decomposed by an infinite series of polynomials represented by
where Wℓ(x,t) will be evaluated recurrently, and Gℓ and Hℓ are the so-called polynomial of W0,W1,...,Wℓ established by [65].
The first few Adomian polynomials are presented as follows:
For ℓ=0,1,2,3,...
The approximate solution for Example 4.4 is expressed as:
Case Ⅱ. Here, we surmise ABC fractional derivative operator coupled with the Jafari transform and Adomian decomposition method. Applying the Jafari transform for Example 4.28.
Taking into consideration the IC given in (4.29), we have
Employing the inverse Jafari transform, we obtain
Thanks to the JDM, we find
Here, we surmise that the unknown function W(x,t) can be written by an infinite series of the form
Also, the non-linearity Fȷ(W),ȷ=1,2 can be decomposed by an infinite series of polynomials represented by
where Wℓ(x,t) will be evaluated recurrently, and Dℓ and Eℓ are the so-called polynomial of W0,W1,...,Wℓ established defined in (4.32). Then, we have
For ℓ=0,1,2,3,...
The approximate solution for Example 4.4 is expressed as:
For λ=1, we obtained the closed form solution of Example 4.4 as
Figure 7 shows the evolutionary outcomes for the explicit and approximate solutions of Example 4.4 for the particular instance λ=1. The result generated by the proposed technique is remarkably similar to the exact solution, as shown in Figure 7.
Then, by considering only the first few elements of the nonlinear equations features are integrated, we can deduce that we have accomplished a reasonable estimation with the numerical solutions of the problem. It is obvious that by adding additional components to the decomposition series (4.33) and (4.36), the cumulative error can be diminished.
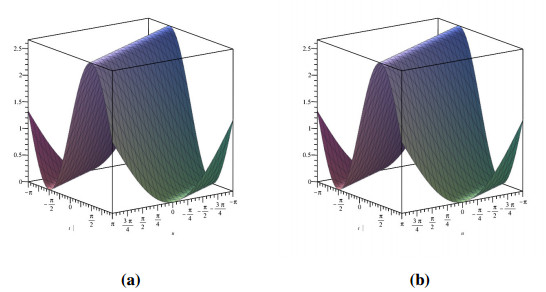
Analogously, we demonstrate the two-dimensional view of the change in fractional values of the order. Figure 8 depicts the response for exact CFD and AB fractional derivative operators.
For a variation of the K(3,3) equation, constrained traveling-wave solutions are achieved. We acquire hump-shaped and valley-shaped solitary-wave solutions, as well as some periodic solutions, for the focusing branch. It is worth noting that optimal focusing provides the aggregate of the frequency and amplitude of the originating waves in the engaging phase, as illustrated in reference [66].
5.
Conclusions
In this paper, we conducted a novel algorithm based on the Jafari transform and Adomin decomposition method, known as the Jafari decomposition method. In the time-fractional technique, we investigated several models such as KdV, mKdV, K (2,2), and K (3,3). To comprehend their physical interpretation, we researched and examined several novel families of solutions and their simulation studies, presented in two-dimensional and three-dimensional plots. The new discoveries concerned the hyperbolic function, trigonometric function, exponential function, and constant function. These new solutions and results might be appreciated in the laser, plasma sciences and wave pattern. To summarise, the suggested method stated above was determined to solve this collection of challenges by utilizing successive fast converging approximations without any limiting requirements or manipulations that changed the physical attributes of the concerns. Also, increasing the recursive procedure leads to the closed form solution of the governing equation.
Conflict of interest
The authors declare that they have no competing interests.









 DownLoad:
DownLoad: