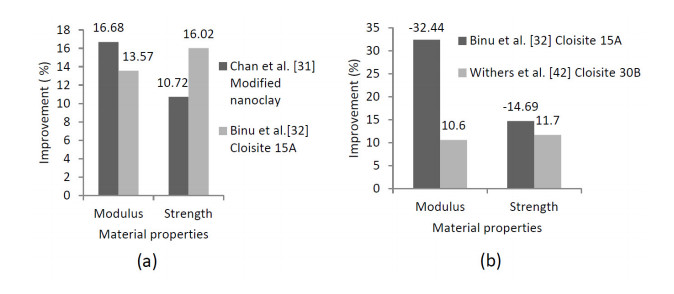
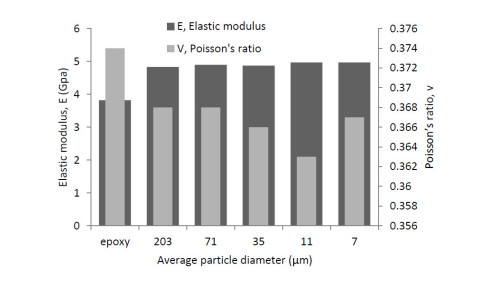
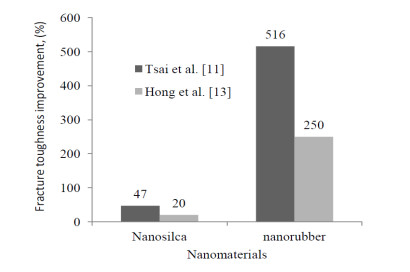
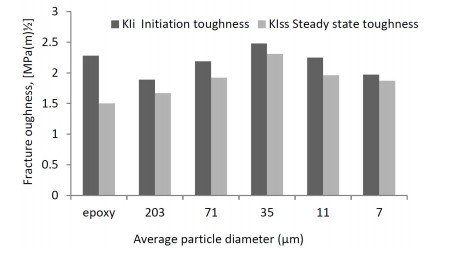
The positive influence of nanomaterials and particles on the behavior of composite material structures have been studied which include the material structural characteristics, manufacturing process, compatibility with the other phases, size, dispersion process, adhesion, etc. The review on the choice of nanomaterials for a specific application and their effects on the bulk materials related to loadings have been overlooked. Thus, this paper reviewed the effects of nanomaterials based on loading conditions, sizes adhesions for the specific category of fillers. It also showed the appropriate filler amount for the enhancement of mechanical properties (i.e. stiffness and strength) and fracture toughness for both interlaminar and intralaminar perspectives. Furthermore, the effects of soft, hard and hybrid fillers were organized to put in evidence how some filler have magnificent effects for specific property enhancements. Moreover, the optimum nanomaterials application related to loading conditions were articulated in order to give a quick suggestion to the structural design engineers and researchers. Finally, the review gives a hint on how the addition of nanofillers and particle affects damage initiations and behavior of fiber reinforced plastic composites.
1.
Introduction
Cancer is considered to be the "number one killer" of human beings and is called an incurable disease [1]. Cancer has become a major enemy that endangers human health and life, and it predicts a poor prognosis of patients with cancer [2]. In recent years, relevant studies have shown that the occurrence and development of cancer are closely related to environmental factors and lifestyle changes [3]. To better understand the progression of cancer, joint efforts are needed in many aspects.
Non-chromosomal structure maintenance protein condensin complex I subunit H (non-SMC condensing I complex subunit H, NCAPH) encoded by a gene located on chromosome 2q11.2, belongs to the bar gene family and a regulatory subunit of the condensin complex [4,5,6,7]. It has been found to be associated with cell proliferation and plays a key role in mitotic chromosome structure and segregation [8,9,10]. Many studies have shown that NCAPH is involved in the evolution of various malignant tumors. Sun et al. found that NCAPH is significantly higher in cancer tissues than in normal non-cancerous tissue, and its high expression is closely correlated with the poor prognosis of patients in hepatocellular carcinoma [11]. It has been reported that high expression of NCAPH promoted the proliferation, migration, and invasion of gastric cancer cells in vitro and in vivo [12]. However, the progression of the specific mechanism of NCAPH in cancer immunology has not been well validated yet.
It is well known that tumor microenvironment (TME) is mainly composed of immune cells including CD8+T cells, CD4+T cells, regulatory T cells, tumor-associated macrophages (TAMs), tumor-associated neutrophils, and natural killer (NK) cells. They interact in the complex TME to regulate tumor development, invasion and metastasis [13,14]. In most cases, the main role of the TME is immunosuppression, which is able to inhibit the activation of lymphocytes in tumor tissue and significantly affect the antitumor effects of drugs. However, current immunotherapy works only in a subset of patients, so we need to find more effective therapeutic targets.
In this study, we accessed the TCGA and GEO databases to analyze the significance of NCAPH in cancer. Various databases such as TIMER, GSEA, Kaplan-Meier Plotter, etc. have shown that NCAPH has significant clinical correlations with the poor prognosis, DNA repair and replication, protein phosphorylation, immune cell infiltration and immune markers. These results revealed the prognostic value of NCAPH and a clear correlation between NCAPH and immune infiltration in pan-cancer.
2.
Materials and methods
2.1. TIMER database
TIMER2 database (http://timer.cistrome.org/) uses RNA-Seq expression profiling data to detect the infiltration of immune cells in tumor tissues. TIMER provides the infiltration of six types of immune cells (B cells, CD4+T cells, CD8+T cells, Neutrphils, Macrophages and Dendritic cells). We analyzed the different expression of NCAPH in tumor tissues and the correlation with immune infiltration with TIMER algorithm [15].
2.2. GEPIA database
For tumors that are without normal tissues data, the module of GEPIA2 (http://gepia2.cancer-pku.cn/#analysis) was further adopted to analyze the box plots of NCAPH expression in the remaining cancers [16]. GEPIA2 also contains cancers of genotype-tissue expression (GTEx) database, which provides more convincing evidence. Moreover, the GEPIA2 database could also be utilized to retrieve the NCAPH expression in different pathological stages (including stage Ⅰ–Ⅳ).
2.3. UALCAN database
The UALCAN portal (http://ualcan.path.uab.edu/analysis-prot.html) was intended to analyze protein expression by taking advantage of data from clinical proteomic tumor analysis consortium (CPTAC) [17]. We obtained available datasets of multiple tumors to explore the total protein expression of NCAPH, including its expression in liver, lung, cervical, glioma, and pancreatic cancers.
2.4. Kaplan-Meier plotter database analysis
GEPIA2 database was performed to generate the survival significance map data of NCAPH more than 30 different forms of cancer, consisting of RFS (Recurrence Free Survival), DMFS (Distant Metastasis Free Survival), PPS (Post Progression Survival), PFS (Progression Free Survival), DSS (Disease Specific Survival), and FP (Full Period of Service). We also obtained the survival plots through the Kaplan-Meier plotter database, which utilized the log-rank test for hypothesis test.
2.5. Genetic alteration database analysis
The cBioPortal (http://www.cbioportal.org) web was applied to explore the mutation frequency, copy number alteration and mutation type of NCAPH [18]. We also obtained the detailed genetic alteration information in the mutations section. In addition, database searches also uncovered associations of NCAPH gene alterations with specific cancer prognoses (OS, DSS, DFS, RFS, and PFS).
2.6. PhosphoSite Plus database analysis
PhosphoSite Plus (https://www.phosphosite.org) website supplied comprehensive information support to study on protein post-translational modifications (PTMs) [19]. Using this website, we obtained the phosphorylation features of the NCAPH protein. The diversity of phosphorylation alteration of NCAPH protein in tumor and normal tissues was further analyzed via the UALCAN portal. The expression and localization of human proteins in different tissues and organs can be retrieved through HPA (https://www.proteinatlas.org) [20]. Furthermore, HPA detected NCAPH expression in a number of different cancer types.
2.7. Data acquisition
The TIMER1 database (https://cistrome.shinyapps.io/timer/) was originally a database used to retrieve tumor-infiltrating immune cell (TIIC) abundance in all TCGA tumors, and it was the first version of TIMER2 database [21]. Herein, the TIMER1 database was used to analyze the relevance of NCAPH and TIICs in different cancers. Then, the TIMER2 database further performed to re-evaluate the relationship among three TIICs and NCAPH expression in the overall cancer level.
2.8. Protein interaction networks
The STRING (https://string-db.org/) was defined to display protein-protein interaction networks, and can be used for functional enrichment analysis [22]. Therefore, we first retrieved the top 50 proteins with potential binding to NCAPH via the site. Subsequently, we identified the top 100 targeted genes that may be related to NCAPH from the "Similar Gene Detection" module of GEPIA2. Jvenn supported intersection analysis and produce Venn diagrams [23].
2.9. Bioinformatics
We performed gene ontology (GO) enrichment and kyoto encyclopedia of genes and genomes (KEGG) pathway analysis through bioinformatics resources web server applied for annotation, visualization and integrated discovery (DAVID) [24,25]. Functional annotation charts of KEGG pathway and GO enrichment analysis which that included biological process (BP), cellular component (CC), and molecular function (MF), were plotted using bioinformatics, freely available online. (http://www.bioinformatics.com.cn) [26].
2.10. Statistical analysis
Data changes on NCAPH were obtained through the cBioPortal website, and data related to its mRNA expression were retrieved. The prognostic significance of each variable in the articles was assessed by Kaplan-Meier survival curves and compared using the log-rank test [27]. Meanwhile, Spearman's correlation test was used for correlation analysis. Unless otherwise specified (P < 0.05), the difference was statistically significant.
3.
Results
3.1. Expression of NCAPH in human pan-cancer
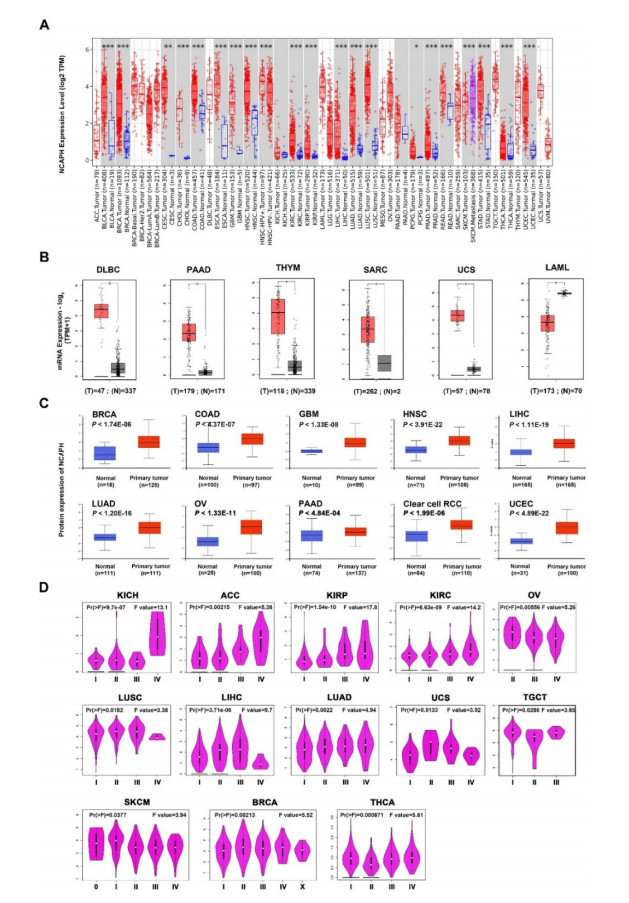
Difference in expression of NCAPH between human cancer and normal tissues was discovered via TIMER2 database. According to Figure 1A, NCAPH in tumor tissues was obviously higher than that of normal tissues, including bladder urothelial carcinoma (BLCA), breast invasive carcinoma (BRCA), cholangiocarcinoma (CHOL), colon adenocarcinoma (COAD), esophageal carcinoma (ESCA), glioblastoma multiforme (GBM), head and neck squamous cell carcinoma (HNSC), kidney chromophobe (KICH), kidney renal clear cell carcinoma (KIRC), kidney renal papillary cell carcinoma (KIRP), liver hepatocellular carcinoma (LIHC), lung adenocarcinoma (LUAD), lung squamous cell carcinoma (LUSC), pancreatic adenocarcinoma (PAAD), prostate adenocarcinoma (PRAD), rectum adenocarcinoma (READ), stomach adenocarcinoma (STAD), uterine corpus endometrial carcinoma (UCEC) (P < 0.05), cervical squamous cell carcinoma and endocervical adenocarcinoma (CESC), and pheochromocytoma and paraganglioma (PCPG) (P < 0.01), but it was no markedly change in thyroid carcinoma (THCA). Similarly, the GEPIA2 database was searched for NCAPH mRNA expression in pan-cancer tissues. It was up-regulated significantly in lymphoid neoplasm diffuse large B-cell lymphoma (DLBC), PAAD, thymoma (THYM), sarcoma (SARC) and uterine carcinosarcoma (UCS) compared with that of normal tissues, while down-regulated in acute myeloid leukemia (LAML) (Figure 1B, P < 0.05).
Consistent with the results, we found that the total protein level of NCAPH was markedly elevated in primary tissues than in normal tissues based on the CPTAC dataset (Figure 1C, P < 0.001). The correlation between NCAPH and the clinical stage of various tumors suggested that the expression of NCAPH was related to the clinical stage of certain cancers, consisting of KICH, adrenocortical carcinoma (ACC), KIRC, KIPR, OV, LUSC, LIHC, LUAD, UCS, TGCT, SKCM, BRCA and thyroid carcinoma (THCA). In contrast, the expression of NCAPH in clinical stage Ⅳ was lower in LUSC and LIHC than in other stages (Figure 1D, P < 0.05). These data suggested that NCAPH expression was generally overexpression in cancer tissues, and it might be involved in the malignant progression of tumors.
3.2. High NCAPH expression predicts the poor prognosis in pan-cancer
The prognostic significance of NCAPH expression in different tumor patients was evaluated using the TCGA and GEO datasets. It turned out that the OS of patients with NCAPH overexpression was significantly shorter than patients in the low expression group, covering ACC, KIRC, KIRP, brain lower grade glioma (LGG), LIHC, LUAD, MESO, PAAD, SARC and uveal melanoma (UVM) (P < 0.05) (Figure 2A). At the same time, NCAPH overexpression was also turned out to reduce DFS in ACC, KICH, KIRC, KIRP, LGG, LIHC, PAAD, PRAD, SARC, TGCT, THCA and UVM (P < 0.05) (Figure 2B). We further found that the overexpression of NCAPH could predict unfavorable outcome in Breast Cancer (P < 0.01), Liver Cancer (P < 0.01), Lung Cancer (P < 0.01) and Ovarian Cancer (P < 0.05) through Kaplan-Meier plotter database (P < 0.05). While the overexpression of NCAPH showed patients with gastric cancer have a longer survival period, the specific molecular mechanism needs to be further studied (P < 0.05) (Figure 3). Thus, the results indicated that overexpression of NCAPH in most tumors predicts a short survival in patients.
3.3. NCAPH-related partners enrichment analysis data
To further elucidate the potential molecular mechanism of NCAPH in tumorigenesis, NCAPH-related genes were screened for biological function analysis. Based on STRING, we retrieved the top 50 NCAPH-binding proteins validated by experimental data, and exhibited their PPI network (Figure 4A). Figure 4B showed that NCAPH expression was notably related to cell proliferation-related genes (BUB1, R = 0.84; CCNB2, R = 0.82; CDCA5, R = 0.82) and cell cycle regulation genes (KIF2C, R = 0.84; KIFC1, R = 0.81; TPX2, R = 0.81) of the top 100 genes (P < 0.01). We also illustrated that NCAPH was positively related to the six genes (BUB1, CCNB, CDCA, KIF2C, KIFC1 and TPX2) in most types of cancer (Figure 4C). Further searching through the Venn diagram database revealed that a total of 29 genes overlapped with each other in the two groups, they are closely related to proliferation function and cell cycle regulation function (Figure 4D).
We then performed KEGG and GO enrichment analysis by combining these two datasets, which contain a total of NCAPH-related genes. The KEGG analysis revealed that these genes were primarily concentrated in ways related to DNA replication and cell cycle signaling pathway (Figure 4E). Interestingly, it was also related to the PD-L1 checkpoint pathway and the inflammatory pathway NF-kB, suggesting that there was a certain relationship between NCAPH and the immune process. GO analysis revealed that the major BP contained cell division and DNA repair (Figure 4F). From the distribution of CC, most of the CC terms were associated with the nucleoplasm (Figure 4G). For the analysis of MF (Figure 4H), it was revealed that NCAPH was mainly related to protein binding. Consistent with our previous gene association findings, these results further suggested that NCAPH may be involved in molecular mechanisms regulating cell cycle and immune escape.
3.4. NCAPH expression is correlated with immune infiltration level in pan-cancer
With the previous results (Figure 1), the mRNA and total protein of NCAPH were overexpressed in BRCA, COAD and other cancers. The TIMER database was further selected to investigate the potential association between the expression level of NCAPH and the level of tumor-infiltrating immune cells (TIICs) in diverse cancer types. The present database showed that NCAPH expression in THCA and KIRC was positively correlated with B cells, CD8+T cells, CD4+T cells, macrophages, neutrophils and DCs (P < 0.05). In contrast, NCAPH expression was significantly negatively correlated with macrophages (r = −0.014, P = 6.68e−01) in BRCA, CD8+T cells (r = −0.129, P = 4.56e−03) in OV, CD4+T cells (r = −0.143, P = 6.32e−02) in PAAD, and neutrophils (r = −0.122, P = 1.96e−01) in THYM (Figure 5A). Figure 5B exhibited the correlation of NCAPH with the level of immune cell infiltration in other types of cancer based on sangerbox. As shown in supplementary Figures S1 and S2, which demonstrates the correlation between NCAPH and the level of immune infiltration in a variety of cancers. Next, the potential relationship between NCAPH expression and immune cell infiltration in different TCGA tumors was considered. The results displayed that NCAPH expression and CD8+T cell infiltration showed a positive correlation in KIRP, LUAD, BRCA and THYM, while negatively correlated in HNSC (HPV-) and UCEC cancers. Consistently, we observed a positive relationship between NCAPH expression and myeloid dendritic infiltration in BRCA and THYM (Figure 6A, B, P < 0.05).
Correspondingly, there was also a clear correlation between NCAPH expression levels and immune marker genes in BRCA (Figure 6C). The results suggested that in BRCA, there was a positive association with NCAPH expression in M2 (ARG1, MRC1, both P < 0.05), TAM (CD80, CD86, both P < 0.05) and B cell (CD38, P < 0.05). In particular, the expression of NCAPH was most significantly correlated with CD80 (P = 5.67e-39, R = 0.379). Moreover, Figure S3 also showed the correlation between NCAPH and immune infiltration markers in other tumors. Therefore, NCAPH may play an immunosuppressive role in tumors through CD80 and assist tumor immune escape.
3.5. Mutation and protein phosphorylation modification of NCAPH in cancers
To identify the mechanism by which NCAPH impacts survival, we used cBioPortal to explore alteration frequencies, including mutation, structural variant, amplification, deep deletion, and multiple alterations, of NCAPH in different cancer types (Figure 7A). Results showed that the top five cancer types with more total mutations were UCEC (5.1%), SKCM (3.38%), STAD (3.18%), KICH (3.08%), and LUAD (2.12%). For specific alteration types, amplifications of NCAPH were enriched in UCS (3.5%), BLCA (1.7%), LUSC (1.64%), HNSC (0.96%), and LUAD (0.88%). We also found one NCAPH-deep deletion was enriched in DLBC (2.08%). At the same time, missense mutation was the commonest mutational style of NCAPH gene in TCGA tumors, and D469lfs/Rfs alteration in the Cnd2 bind domain was discovered in COAD, UCEC and STAD (Figure 7B). Based on these results, we further studied the correlation of NCAPH alteration with prognosis in the top five cancer types and found the prognostic value of NCAPH alteration. The results are summarized in Figure 7C. Altered NCAPH is significantly associated with a good prognosis in HNSC (OS (p = 0.0408), PFS (p = 0.0137), DSS (p = 0.0499)) and BLCA (DSS (p = 0.0229)). In addition, NCAPH has different protein phosphorylation sites in HNSC, and its expression in tumors was also different (P < 0.001). Higher NCAPH phosphorylation level was attended in HNSC (S59, S67, S76, S190, S222 and T38, P < 0.05). Conversely, NCAPH phosphorylated S16 was downregulated in tumor tissues, with no significant difference in phosphorylated S14 (Figure 7D).
4.
Discussion
It has been reported that NCAPH is a mitosis-associated protein that plays an important role in tumorigenesis and tumor prognosis assessment [28]. However, it was unclear whether NCAPH has a common pathway in different tumor pathogenesis. We also failed to retrieve any literature or report about NCAPH in pan-cancer. Thus, the objective was to evaluate the prognostic value and the latent biological functions of NCAPH in different types of cancer.
Our analysis demonstrated that NCAPH was highly expressed in most cancers compared with normal controls and positively correlated with the clinical stage. Interestingly, NCAPH overexpression in clinical stage Ⅳ was lower in LUSC and LIHC, which may be related to factors such as cancer type and the number of patients in the database, which further leads to the heterogeneity of NCAPH function. Likewise, the aberrant NCAPH expression usually indicates an adverse prognosis for many kinds of cancer, which may serve as a prognostic biomarker in cancer patients. Lu et al. found that NCAPH protein was highly expressed in breast cancer, and its overexpression predicts poor prognosis in patients [29]. Cui et al. demonstrated that the OS was lower in prostate cancer patients with high NCAPH expression [30]. This finding was in line with our previous analysis. Whereas, we still need to fully consider other clinical features and provide more direct evidence to confirm whether the over- expression of NCAPH was involved in the regulation of tumor malignant evolution, or was merely an outcome of anti-tumor changes in normal tissue. Together, these results strongly indicated that NCAPH may have value as a prognostic indicator for cancer.
Genetic mutations acted as a significant player in the development and growth of cancer, and they can also be targets for effective treatments [31,32]. It has been reported that the NCAPH gene with a high mutation rate in cancer, and it has an aggressive phenotype [33]. Meanwhile, our research identified that missense mutation was the most frequent mutational style of NCAPH gene in TCGA tumors. Noteworthy, NCAPH gene mutations were significantly enriched in STAD, the overexpression of NCAPH in gastric cancer predicts a longer survival of patients. Therefore, we conjecture that the mutation of NCAPH at the gene locus may be closely related to the longer survival of patients. Studies had reported that phosphorylated NCAPH in multiple tumors was well correlated with tumorigenesis [34,35,36]. The phosphorylation of NCAPH at ser988 had been found to promote the occurrence of BRCA1 [37]. Herein, our results noted that NCAPH phosphorylation (S59, S67, S76, S90, S222 and S38) is higher in HNSCs, but S16 is lower in HNSCs. Whether these post-translational modification sites have clinical significance remains unclear, and further clarification was required by subsequent molecular biology experiments.
Recently, multiple related reports have obtained that protein gene mutations and protein phosphorylation modifications also affect the level of tumor immune infiltration in different cancer types [38,39,40]. Our study elucidated the correlation between NCAPH expression and immune cell infiltration levels in various tumors, especially CD8+T cells, DCs and neutrophils. Consistent with our research, Yin et al. proposed that NCAPH overexpression is associated with poor prognosis and immune infiltration in COAD [41]. Moreover, macrophages as the first line of immune defense play an important role in every stage of tumor development and are the core regulators in the TME [42]. Interestingly, NCAPH was mainly positively correlated with M2 macrophage markers, such as CD80. Meanwhile, in our study, NCAPH expression was significantly enriched in PD-L1/NF-kB pathway, DNA replication and DNA repair pathways. The discovery that the NCAPH has plays an important role in cancer immunology and facilitates future studies with larger patient populations that could determine the feasibility of clinically integrated therapy.
In summary, NCAPH is correlated with the poor prognosis of patients, protein phosphorylation, immune cell infiltration, and immunity markers in multiple cancers. NCAPH may serve as a pan-cancer prognostic biomarker and play a key role in cancer as a new immunotherapeutic target. However, there is still a shortcoming in the molecular mechanism of NCAPH regulating tumors, and the specific cell function experiments need to be further explored. It is worth noting that although we have found NCAPH in multiple online datasets as a potential cancer biomarker and associated with poor prognosis for multiple cancer patients. But so far there is no report on the expression of NCAPH in BLCA, UCEC, GBM, CHOL, etc. This requires us to carry out more studies to improve the significance of the differential expression of NCAPH in different types of cancer in prognostic evaluation.
Acknowledgments
This research was supported by the Project from the Department of Health of Jilin Province (2020J002). The National Natural Science Foundation of China (No. 82160552).
Conflict of interest
The authors declare that there is no conflict of interest.









 DownLoad:
DownLoad: