1. Introduction
Proteostasis, or protein homeostasis, controls proteome by regulating mRNA targeting, proper protein synthesis, folding, trafficking and degradation [1], all necessary processes to keep cell functionality [2].
The processes in the cell involved in protein homeostasis can be grouped in those contributing to the synthesis of new proteins (mRNA processing, transport and translation) and those involving protein degradation or removal (chiefly, the ubiquitin-proteasome system and the autophagy-lysosome system). Other cellular mechanisms described to respond to protein unbalance or misfolding, as those encompassing cellular stress responses including the endoplasmic reticulum unfolded protein response [3], will not be addressed in this review.
In the neuronal context, proteostasis mechanisms are intimately associated to brain function. Notably, some forms of autism have been associated to the deregulation of proteostasis [4]. Neurons are polarized cells with long dendritic and axonal projections that receive information through highly specialized subcellular compartments called synapses. One neuron may contain around 10,000 to 30,000 of them. Synapses may be located close, or certainly far from the cell body, meaning that neurons need mechanisms that allow those synapses to function, to some degree, in an autonomous way, but coordinated with the cell body, to respond to local activity as well as to local signaling cues. This is partially supported by the segregation of axonal and somatodendritic membrane micro-domains that limit the diffusion of specific membrane components [5]. In addition, spatially-segregated protein synthesis contributes to the maintenance of neuronal compartments functionally differentiated [6]. We will focus in this review on those mechanisms characterized at the synaptic level. When the presynaptic terminal sends a signal to a specific postsynaptic terminal across the synaptic cleft, the postsynaptic terminal undergoes activity-dependent protein composition changes. These local protein modifications in abundance and activity are the bases supporting synaptic plasticity, thus altering the characteristics of that exact synapse. Those plasticity mechanisms might turn the synapse more efficiently coupled to the presynaptic signal, through potentiation processes (short-term, or long-term potentiation, depending on their duration), or may reduce the coupling between presynaptic and postsynaptic sides, through depression processes (short-term or long-term depression) [7,8]. This plasticity can be bidirectional, since postsynaptic terminals may produce retrograde diffusible messengers to affect presynaptic activity. This is, for example, the case of nitric oxide [9] or endocannabinoids [10] that affect presynaptic function.
To achieve activity-dependent protein composition changes in response to input signals, rapid alterations in local protein synthesis and function are necessary [11,12]. In fact, synaptic plasticity implies morphological and functional changes controlled by the spatial restriction of protein translation [13,14] and protein degradation [15]. Thus, local proteostasis determines proper plasticity in synapses.
Proteostasis deregulation underlies some forms of autism spectrum disorder (ASD) [16]. These are neurodevelopmental disorders characterized by the impairment of the child's ability to communicate and interact with others, and the appearance of restricted repetitive behaviors causing a wide range of social or occupational dysfunction [17]. The etiology of most forms of autism is unknown, although there is a clear genetic association [18,19]. For those cases of ASD with an identified etiology, it has been observed that in many occasions those genes affected are involved in synaptic protein homeostasis (Table 1). Interestingly, the deregulation of protein homeostasis found in a number of ASDs is largely associated to an intracellular signaling pathway, the mechanistic/mammalian target of rapamycin (mTOR) pathway. This signaling pathway is known to support synaptic plasticity in the brain by controlling protein synthesis and degradation [15,20,21]. The present review is particularly focused on those synaptic processes in proteostasis where mTOR signaling seems to play a relevant role given the characteristics of the disorders associated to its dysfunction.
Table 1. Summary of autism susceptibility genes involved in proteostasis.
| Gene |
Disorders |
Function |
Reference
|
| CELF1/CUG-BP1 |
Myotonic dystrophy, type 1 |
RNA binding protein |
[163]
|
| DISC1 |
ASD/Asperger syndrome, Schizophrenia (SCZ) |
Multifunctional interacting protein |
[164,165]
|
| ELAVL3 |
ASD |
RNA binding protein |
[166,167]
|
| FMR1 |
ASD, Attention Deficit Hyperactivity Disorder (ADHD), Developmental Delay (DD), Epilepsy (EP) Intellectual Disability (ID) |
RNA binding protein |
[168,169,170]
|
| RBFOX1 |
ASD, DD, EP, ID |
RNA binding protein |
[171,172,173]
|
| RNPS1 |
ASD, DD, ID |
RNA binding protein |
[174]
|
| SNRPN |
ASD |
Tissue-specific alternative RNA processing |
[175,176]
|
| MECP2 |
ASD, ADHD, DD, EP, ID, SCZ |
Methylation-dependent transcriptional repression activity, RNA processing |
[177,178,179]
|
| DOLK |
ASD, EP, ID |
Dolichol kinase, glycosylation |
[180]
|
| PTEN |
ASD, ADHD, DD, EP, ID |
Phosphatase: mTOR negative regulator via PI3K |
[181,135,182]
|
| NF1 |
ASD |
Ras GTPase: Ras-MAPK negative regulator |
[183,126]
|
| TSC1 |
ASD, DD, ID |
GTPase activator protein: mTOR negative regulator via Rheb |
[168,184]
|
| TSC2 |
ASD, DD, EP, ID |
GTPase activator protein: mTOR negative regulator via Rheb |
[121,184,185]
|
| CUL3 |
ASD, SCZ |
E3-ubiquitin ligase |
[186,187]
|
| CUL7 |
ASD |
E3-ubiquitin ligase |
[166,167]
|
| HECW2 |
ASD |
E3-ubiquitin ligase |
[166,167]
|
| HERC2 |
ASD, DD, ID |
E3-ubiquitin ligase |
[188,189]
|
| HUWE1 |
ASD, DD, ID |
E3-ubiquitin ligase |
[190,191]
|
| RNF135 |
ASD |
E2-dependent E3-ubiquitin ligase |
[192]
|
| UBE2H |
ASD |
Ubiquitin ligase |
[193]
|
| UBE3A |
ASD, DD, EP, ID |
E3-ubiquitin ligase |
[194,195]
|
| UBE3B |
ASD, DD, ID |
E3-ubiquitin ligase |
[196,197]
|
| UBE3C |
ASD |
E3-ubiquitin ligase |
[187]
|
| UBL7 |
ASD, ID |
Ubiquitin binding |
[198]
|
| UBR5 |
ASD, EP |
E3-ubiquitin ligase |
[199,167]
|
| UBR7 |
ASD |
E3-ubiquitin ligase |
[200]
|
| USP7 |
ASD, DD, ID |
Deubiquitination |
[201,166]
|
| USP9Y |
ASD |
Polyubiquitin hydrolase |
[202]
|
| PSMD10 |
ASD, SCZ |
Non-ATPase proteasome subunit of the 19S regulator: protein degradation |
[203]
|
| PYHIN1 |
ASD |
Transcriptional regulation |
[204,167]
|
| CAPN12 |
ASD |
Calcium-regulated non-lysosomal thiol-protease |
[166,205]
|
| DPP4 |
ASD |
Serine exopeptidase |
[204,206]
|
| DPP6 |
ASD, ADHD, ID, TS |
Promotes cell surface expression of the KCND2 potassium channel |
[207,208,209]
|
| DPP10 |
ASD |
Promotes cell surface expression of the KCND2 potassium channel |
[171,208]
|
| Gene code and corresponding protein name, CELF1/CUG-BP1: CUG triple repeat RNA binding protein 1; DISC1: disrupted in schizophrenia 1 protein; ELAVL3: ELAV-like protein 3; FMR1: fragile X mental retardation protein; RBFOX1: RNA binding protein fox-1 homolog; RNPS1: RNA binding protein with serine-rich domain 1; SNRPN: small nuclear ribonucleoprotein polypeptide N; MECP2: methyl-CpG-binding protein 2; DOLK: dolichol kinase; PTEN: phosphatase and tensin homolog; NF1: neurofibromin; TSC1/2: tuberous sclerosis complex 1/2; CUL3: cullin-3; CUL7: cullin-7; HECW2: HECT, C2 and WW domain containing E3 ubiquitin protein ligase 2; HERC2: HECT and RLD domain containing E3 ubiquitin protein ligase 2; HUWE1: HECT, UBA and WWE domain containing 1, E3 ubiquitin protein ligase; RNF135: ring finger protein 135; UBE2H: ubiquitin conjugating enzyme E2 H; UBE3A: ubiquitin protein ligase E3A; UBE3B: ubiquitin protein ligase E3B;UBE3C: ubiquitin protein ligase E3C; UBL7: ubiquitin-like 7; UBR5: ubiquitin protein ligase E3 component N-recognin 5; UBR7: ubiquitin protein ligase E3 component N-recognin 7; USP7: ubiquitin specific peptidase 7; USP9Y: ubiquitin specific peptidase 9, Y-linked; PSMD10: proteasome 26S subunit, non-ATPase 10; PYHIN1: Pyrin and HIN domain family member 1; CAPN12: calpain 12; DPP4: dypeptidyl peptidase like 4; DPP6: dypeptidyl peptidase like 6; DPP10: dypeptidyl peptidase like 10. |
2. Proteostasis Mechanisms in Synaptic Function
As mentioned above, synaptic proteostasis will depend on cellular processes resulting in the synthesis of new proteins, or removing pre-existing ones, directed by synaptic activity triggered by the surrounding stimuli.
2.1. Synaptic targeting and expression modulation of mRNAs
2.1.1. mRNA processing and transport
Newly synthesized mRNAs are transported to translation sites outside the nucleus. The regulation of this process is crucial for synaptic protein homeostasis and plasticity [22]. mRNA molecules have been found on dendrites close to the synapse to support, through controlled translation, synapse plasticity in a stimulus-dependent fashion [23]. In this way, swift changes in protein composition may rapidly respond to neighboring stimuli in a spatially and temporally restricted manner [24,25]. Notably, mRNA local translation regulation is one of the crucial processes supporting the synaptic tagging and capture hypothesis [26]. This hypothesis explains those synaptic alterations necessary discriminate specific synapses in the context of the formation of lasting memories. Thus, stimulated synapses are first tagged by activity-derived modifications, which subsequently capture plasticity related proteins/particles (PRPs), synthesized after synaptic stimulation, allowing plasticity, and therefore memory consolidation-prone modifications, in those previously tagged synapses [26,27]. Hence, the synaptic tagging and capture hypothesis incorporates those cellular processes relevant for mRNA transport and local translation regulation (see below) [28,29].
mRNA molecules may have different elements that support their specific targeting and modulation at synapses. Localization elements or molecular zipcodes are sequence and structural cis- elements in mRNA molecules that determine their localization. In general, zipcodes are mostly found in 3' UTR, and less frequently found in 5' UTR. The targeting of mRNAs to dendrites requires dendritic targeting elements (DTEs) to be bound by trans-acting RNA-binding proteins (RBPs). DTEs have been detected in CaMKIIα [30], β-actin [31], MAP2 [32], Arc [33] and BDNF [34].
Another level of proteostasis control at the synapses is mediated by local RNA splicing. It has been described the presence of spliceosomes in synaptic terminals, so it is thought that regional splicing is also a point of protein translation regulation in synapses [35]. Moreover, it has been proved that several RBPs, such as Sam68, are implicated in splicing [36,37]. In addition, there are mRNAs containing zipcodes in intronic regions [38]. These are known as cytoplasmic intron-sequence retaining transcripts (CIRT), which have been shown to be abundant in brain mRNAs, targeting them to dendrites [39]. Altogether, mRNA transport into dendritic compartments is required to support postsynaptic stimuli-dependent plasticity [24,25].
The localization elements in mRNA molecules are recognized by specific RBPs, which then are bound by other accessory proteins, creating messenger ribonucleoprotein (mRNP) granules [40]. At that point, these macrocomplexes are responsible of mRNA protection from nucleases, its cellular transport and its local translational regulation [25,41]. The mRNP granule transport to the dendrite of destination is carried out by microtubules [42,43]. To this end, mRNP granules have several crucial elements: RBPs, which are in charge of preventing translation before delivering, adaptors to cytoskeletal machinery and molecular motors [44].
2.1.2. mRNA translation control by RNA binding proteins
After mRNP granules arrive to dendrites, mRNA's translation must be regulated so that proteostasis is preserved. RBPs attached to the mRNA molecules play a key role at this point, acting as repressors or promoters of translation [4,28]. Depending on the local synaptic stimulation, the RBPs attached to the mRNA molecule critically determine whether the attached mRNA is translated in order to support long-lasting forms of synaptic plasticity or not [45]. Once the synaptic input arrives, it takes place a signaling cascade that ends with the modification of RBPs. Consequently, the RBPs' affinity to its mRNA cargo is changed, thus regulating the translation of the mRNA molecule [45]. For instance, FMRP1 (fragile mental retardation protein), ZBP1 (zipcode-binding protein) or CPEBs (cytoplasmic polyadenylation element binding proteins) are RBPs that attached to an mRNA molecule function as translation repressors [46,47,48], whereas Sam68 promotes mRNA translation when bound to it [49,50].
The case of β-actin is useful to illustrate mRNA translation regulation by RBPs in dendrites. β-actin mRNA is linked to ZBP1 [51] and Sam68 [52] at the same time. When an input signal arrives, ZBP1 might be phosphorylated. This phosphorylation lowers ZBP1, but not Sam68, affinity to β-actin mRNA, allowing its translation, which is further enhanced by bound Sam68 [47,49]. In the case of CPEBs, CPEB1 to 4 have been described in the brain, and more specifically at the dendrites, where they regulate synaptic plasticity [48]. CPEB1, the founding member of this family, blocks mRNA translation of its target mRNAs by binding to the CPE (cytoplasmic polyadenylation element) present at the 3' UTR. Furthermore, by binding to neuroguidin, it prevents the assembly of the eIF4E-eIF4G components of the translation initiation complex [53,54]. Following activation signals, CPEB1 promotes translation initiation by poly(A) tail elongation and binding of poly(A)-binding proteins (PABPs), which recruit eIF4G to compete with neuroguidin for the binding of eIF4E [55]. Signaling through NMDA (N-methyl-D-aspartate) glutamate receptors present at synapses regulate CPEBs and their target mRNAs: CPEB1 inhibits translation of its target mRNAs until NMDA type-mediated glutamate receptor activation stimulates its phosphorylation by either Aurora kinase A or CaMKIIα, resulting in increased mRNA polyadenylation and translation at synapses [56]; other CPEBs such as CPEB3 shows a different mechanism. CPEB3 must be cleaved by calpain 2 after NMDA glutamate receptor signaling, which results in the translation of CPEB3-targeted/repressed mRNAs such as the AMPA-2 (α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid) glutamate receptor [57].
Several neurological disorders are related to RBPs dysfunction. Among them, FMRP has the characteristics of a RBP [58] and its expression is disrupted in FXS [59]; disrupted in schizophrenia 1 (DISC1), a RBP mutated in this disorder, is vital for dendritic mRNA transport and synaptic plasticity [60]; TDP-43 (transactive response DNA-binding protein 43) regulates splicing, mRNA stability, mRNA transport, translation and synaptic function in motoneurons [61], and it is found deregulated in amyotrophic lateral sclerosis [62]. Interestingly, CPEB1 removal in the context of the mouse model for FXS, a model where FMRP is not expressed, showed that CPEB1 depletion improves the neuronal deficit including affected synaptic plasticity and memory alterations. This genetic rescue suggests that the proteostasis unbalance produced by FMRP absence can be prevented by CPEB1 deficiency [63]. Other cases of mutations affecting RBPs with neurological consequences are detailed in Table 1.
2.1.3. Control of mRNA translation initiation
Once mRNA molecules get to their translation site, in addition to its translational control by bound RBPs and splicing, they must find a supportive environment for translation. These favorable conditions are provided by signaling pathways involved in the regulation of translation initiation, the most limiting step in mRNA translation [64,65]. The process of translation initiation of a mRNA requires the recognition of the cap structure at the 5' end and the recruitment of the ribosome by multiple eukaryotic initiation factors (eIFs). The heteromeric eIF4F complex consists of the cap binding protein eIF4E, the RNA helicase eIF4A, and the protein eIF4G, and all these translation initiation factors are targets of different regulators to finely control protein synthesis [66]. Finally, the translation initiation factor 4B (eIF4B) stimulates eIF4F complex by potentiating the eIF4A RNA helicase activity [66]. Interestingly, the activity of several components of the eIF4F complex is controlled by regulators, such as mTORC1 [67] or MAPK (mitogen‐activated protein kinase) [68] signaling pathways, which stimulates cell translational machinery. eIF4E is the less abundant initiation factor, and its function is sequestered by 4E-binding proteins (4E-BPs), an interaction that is prevented by mTORC1 activity, thus allowing the translation of the mRNA [65]. In addition, eIF4E is phosphorylated by MNK (MAPK interacting protein kinase) activation, which also promotes eIF4F complex activity [69]. The relevance of these mechanisms in the local protein synthesis at the neuronal level is supported by the presence of mTORC1 signaling pathway and eIF4F complex components in dendritic compartments [70]. The mTOR pathway will be further described in the context of ASD below.
2.2. Protein degradation regulation
Degradation regulation is essential to maintain proteostasis in neurons. The ubiquitin-proteasome system and the autophagy-lysosome system are the most relevant proteolytic systems in most cell types [71]. The ubiquitin-proteasome system would be responsible to target misfolded and short-lived proteins, while the autophagy-lysosome system would mediate the degradation of long-lived proteins and organelles [71].
2.2.1. Ubiquitin-proteasome system
The ubiquitin-proteasome system involves the conjugation of several ubiquitin proteins, a 76-amino acid protein, to substrates that must be degraded by the proteasome [72,73]. The poly-ubiquitin chain can be removed or shortened by deubiquitinating enzymes, providing reversibility to the ubiquitination reaction. As a whole, the ubiquitin-proteasome system is highly regulated at all steps [74]. The initial attachment of the poly-ubiquitin chain to the target protein to be degraded occurs through specific enzymatic steps mediated by E3s (ubiquitin ligases), which provides substrate/target specificity. The other enzymes involved are E2s (ubiquitin-conjugating enzymes) and E1 (ubiquitin-activating enzyme) [75]. The normal activity of the ubiquitin-proteasome system is necessary for proper synaptic function [73,76,77]. The proteasome has a relevant role in the synaptic tagging and capture hypothesis of synaptic plasticity [26]. This complex is sequestered in dendritic spines by local synaptic activity [78]. Therefore, protein degradation via proteasome seems essential for the structural and functional changes associated to synaptic plasticity, through the degradation of inhibitory constrains such as translation repressors involved in the establishment of synaptic plasticity [79]. For example, the inhibition of the ubiquitin-proteasome system promotes the accumulation of BDNF (brain-derived neurotrophic factor) creating conditions that potentiate long-term synaptic plasticity [26,80].
Deregulation of the ubiquitin-proteasome system is associated with aging and neurodegenerative diseases [81]. In FXS, there are reports of abnormalities either in the transport of proteasome subunits and ubiquitin ligases (E3) into dendritic spines, or in the activity-dependent ubiquitination of synaptic proteome [82]. Interestingly, there is an interplay between proteasome-mediated protein degradation and protein synthesis control in synaptic plasticity through the mTORC1 pathway [83]. Notably, a number of mutations, in genes encoding for components of the ubiquitin-proteasome system, have been described associated with increased autism susceptibility (Table 1), illustrating the relevance of synaptic protein degradation in neuronal function.
2.2.2. Autophagy-lysosome system
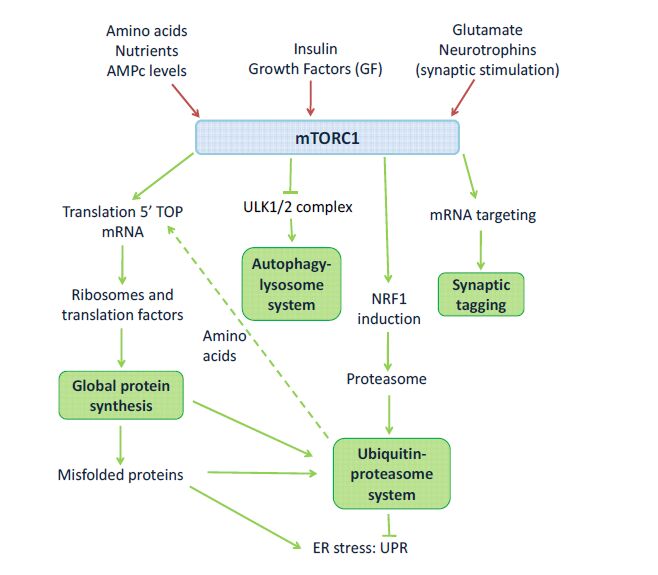
The autophagy-lysosome system manages the transport of cytosolic elements to the lysosome. This transport to lysosome might be chaperone-mediated, directed by the formation of the autophagosome, as is the case of macroautophagy, or directly mediated by the lysosome in a pynocytosis-like event called microautophagy [84]. Autophagosomes merge with lysosomes allowing the degradation of the target content, and contributing to protein homeostasis regulation. Autophagy in neurons is constitutively active and this activity is critical for neuronal survival [85]. The cytosolic elements potentially processed by the autophagy-lysosome system are aged proteins, pathogenic protein aggregates and damaged organelles [86]. The dysfunction of the autophagy-lysosome system is especially relevant to pathological conditions such as neurodegenerative disorders [87], and has been recently associated to ASD [88]. Proper autophagic activity would be relevant during neurodevelopment to perform adequate synaptic pruning, a significant neurodevelopmental process of synapse elimination that occurs between early childhood and the onset of puberty [89,90]. Notably, at the molecular level, mTOR signaling inhibits autophagy(Figure 1) by phosphorylating the ULK1 complexes (UNC-51 like kinase) [87,91]. Therefore, the unbalanced activity of the mTOR signaling seems to be related with the alterations in autophagy and the deficits in spine pruning characteristic of ASDs [88].
In summary, synaptic proteostasis, maintained by different cellular mechanisms described in neurons, involves the synthesis and degradation of proteins driven by synaptic activity and the neuronal context, to support synaptic functionality (Figure 1). Among the signaling pathways involved, the PI3K (phosphoinositide 3-kinase)/mTOR and the ERK/MNK pathways are the most relevant molecular mechanisms implicated in synaptic proteostasis.
3. mTOR Signaling Overview Focus on mTORC1
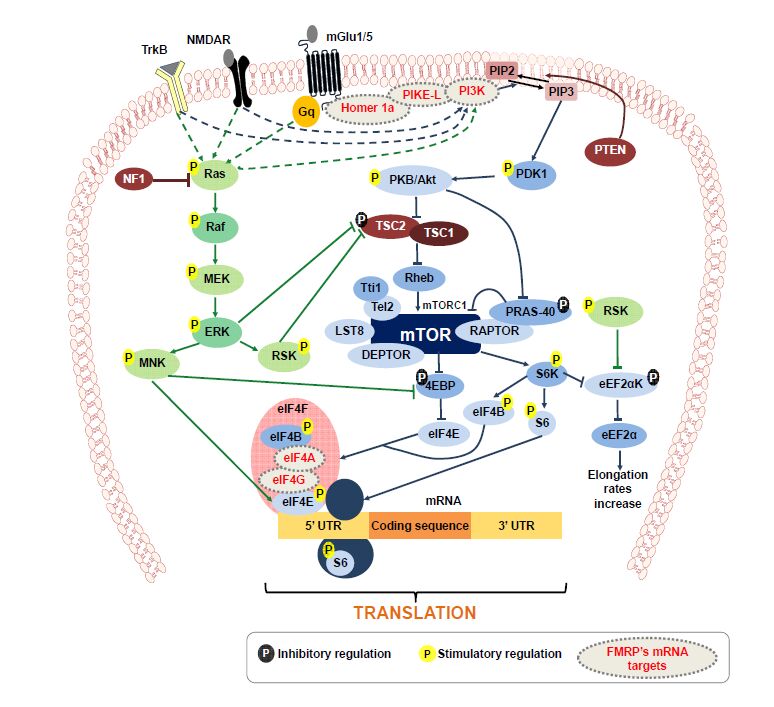
mTOR is a serine/threonine kinase that forms two functionally distinct signaling complexes, mTORC1 (mTOR complex 1) and mTORC2 (mTOR complex 2) [92]. Both complexes share a number of common proteins: mTOR, DEPTOR, LST8/GβL, Tel2 and Tti1 (see Figure 2). In addition, mTORC1 specifically includes RAPTOR and PRAS40. Instead, mTORC2 specifically includes RICTOR and mSIN1. Several specific inhibitors for mTORC1 and dual inhibitors that block mTORC1/mTORC2 activity have been characterized [93], which have allowed to study those specific processes involving mTORC1 or both mTORC1/mTORC2 [21,92]. Unfortunately, no specific inhibitors for mTORC2 have been described so far, and most data on its relevance on brain function comes from genetic targeting of mTORC2 components. Nevertheless, mTORC2 signaling is a vital regulator of actin polymerization [94,95]. In addition, RICTOR conditional deletion in mice revealed reduced mTORC2 activity and impaired long-term memory and long-term plasticity, as well as defective actin polymerization [96]. Complementarily, mTORC2 activity boosting was found to restore memory performance in aged mice [97], indicating a relevant role of this complex in neuronal function that warrants further research.
3.1. mTORC1 as an integrator of neuronal stimuli
In the neuronal context, mTORC1 activation is driven by various extracellular factors such as glutamate [98], BDNF [99] or insulin [100], among others [21,101] (Figure 2). PI3K/PDK1 pathway mediates the activation of PKB/Akt downstream of membrane receptors to modify the activity of tuberous sclerosis complex (TSC), composed by the tumor suppressors TSC1 (hamartin) and TSC2 (tuberin) (Figure 2). TSC can prevent the activation of mTOR by the small GTPase Rheb, a potent activator of mTORC1 when bound to GTP. Importantly, phosphorylation of TSC by PKB/Akt prevents the inhibition of Rheb, leading to mTOR activation (Figure 2). Such mTORC1 activation results in the modulation of downstream effectors with relevance to mRNA targeting and translation, as well as to protein degradation.
mTORC1 activity contributes to synaptic tagging modulating, for example, CaMKIIa mRNA stability and expression with the participation of the RNA-binding protein HuD [102]. At the translational control, mTORC1 phosphorylates S6K, which then phosphorylates S6 ribosomal subunit, eIF4B or eEF2αK. Additionally, mTORC1 phosphorylates 4E-BP at multiple sites, disrupting 4E-BP binding to eIF4E, so the later can bind to the cap structure of the mRNA and to the other components of eIF4F complex to initiate translation [21]. Moreover, mTORC1 activity enhances the translation of 5′terminal oligopyrimidine tract-containing motif mRNAs (5'TOP mRNAs), which are mRNAs coding for ribosomal proteins, elongation factors and translation factors [103]. Together, mTORC1 signaling is involved in a number of key steps leading to mRNA translation at synaptic contacts.
mTORC1 also plays a role in protein degradation. mTORC1 activity results in the induction of the transcription factor NRF1 (also known as NFE2L1), which stimulates the increase in proteasome levels [15] (Figure 1). This extent would facilitate the recycling of amino acids from pre-existing proteins to be used in new protein synthesis. Interestingly, mTORC1 activation inhibits autophagy by phosphorylating the ULK1 complex (91). Therefore, mTORC1 is a signaling node in neuronal function, key in proteostasis with roles at different levels: RNA targeting and stability, mRNA translation, and protein degradation through the ubiquitin-proteasome system and the autophagy-lysosome system (Figure 1).
It is worth mentioning the significant crosstalk between the mTORC1 signaling and MAPK signaling in synaptic proteostasis. Additionally to PI3K-Akt-mTORC1 pathway, neuronal stimuli also trigger Ras-Raf-MEK-ERK signaling pathway, which also plays a major role in increasing global protein translation. This pathway promotes MNK and RSK phosphorylation, both having a role in mTORC1 signaling pathway (Figure 2).
3.2. Syndromic forms of ASD show affected activity of mTORC1
Several autism susceptibility genes encode for proteins involved in proteostasis mechanisms (Table 1). There is direct evidence of the relation of several syndromic forms of ASD with alterations in the mTORC1 signaling pathway: tuberous sclerosis (TS) [104], PTEN-related disorders [105], fragile X syndrome (FXS) [106,107], MECP2 alterations (Rett syndrome [108] and MECP2 duplication syndrome [109]) and Angelman syndrome (AS) [110]. In addition, neurofibromatosis type 1 (NF1) shows an over activation of Ras-Raf-MEK-ERK signaling [111]. These disorders share synaptic alterations pointing to underlying common pathogenic processes [112]. The overstimulation of the mTORC1 cascade can be induced by direct alterations in a mTORC1 pathway component or by alterations in distant regulatory proteins [113]. This situation is associated to increased protein synthesis rates, which may underlie aberrant synaptic plasticity that characterizes the models of these disorders [112,114]. Indeed, this pathway, which has been pinpointed to have a relevant role in structural and functional synaptic plasticity [20], was found crucial in neuronal circuit development [88], most probably due to the close control it exerts over autophagy and protein synthesis in synapses [21]. Interestingly, determinations in human brain tissue from ASD patients show a higher mTORC1 activity than in control tissue, paralleled by a reduction in synapse elimination during neurodevelopment [88]. The alterations in dendritic spine density would be due to developmental synaptic pruning deficits in ASD patients. Synaptic pruning, normally performed by the autophagy-lysosome system would be abnormally inhibited in ASD preventing proper synapse elimination [88] (Figure 1). Interestingly, animal models of some of these disorders improved their neurological deficits by pharmacological inhibition with the mTORC1-specific inhibitor rapamycin or other rapamycin-like inhibitors (Table 2). This is the case for TS [104], PTEN-related disorders [115], FXS [116] and AS [117]. Furthermore, the activation of neuronal autophagy recovers synaptic function and reduces autistic-like behaviors in ASD mouse models with overstimulation of mTORC1 [88].
3.2.1. Tuberous sclerosis
Tuberous sclerosis (TS) is a genetic multisystem disorder characterized by the tumorous growth or malformations (hamartomas) in skin, kidney, lung, heart, liver and brain. The central nervous system manifestations include epilepsy, intellectual disability and ASD. It is caused by heterozygous mutations in either TSC1 [118] or TSC2 [119], components of the TSC complex, where 30% of the cases are familial with autosomal dominant pattern of inheritance, and 70% of the cases are caused by de novo mutations. TSC functions as a GTP-ase activator protein for Rheb [120] (Figure 2). Since TSC inhibits mTORC1 activity, loss-of-function mutations in these proteins lead to mTORC1 TSC1/TSC2-dependent derepression (Figure 2). Several studies, at the clinical and preclinical level, suggest that mTORC1 inhibitors such as rapamycin, RAD001 and everolimus, might be useful to treat the neuronal phenotype (Table 2) of TS [121,122,123] pointing to mTORC1 hyperactivity inhibition as a valuable therapeutic approach in TS.
Table 2. Summary of mTOR pathway changes in animal models of ASD.
| Disorder |
Gene mutated |
mTOR pathway activity |
Sensitive to treatment |
Reference
|
| Tuberous sclerosis |
Tsc1 HZ in neurons |
Enhanced (↑p-S6) |
Rapamycin |
[122]
|
| Tuberous sclerosis |
Tsc2 HZ |
Enhanced (↑p-S6K) |
Rapamycin |
[123]
|
| PTEN-related disorders |
Pten in neurons |
Enhanced (↑p-S6) |
|
[105]
|
| PTEN-related disorders |
Pten in neurons |
Enhanced (↑p-S6) |
Rapamycin
|
[115]
|
| Fragile X syndrome |
Fmr1 KO |
Enhanced (↑p-mTOR, ↑p-S6K, ↑p-S6) |
|
[213]
|
| Fragile X syndrome |
Fmr1 KO |
Enhanced (↑p-S6K) |
Temsirolimus
|
[214]
|
| Rett syndrome |
Mecp2 KO |
Reduced (↓p-mTOR, ↓p-S6K, ↓p-S6) |
|
[108]
|
| MECP2 duplication syndrome |
Mecp2 duplication |
Enhanced (↑p-S6K) |
|
[151]
|
| Angelman syndrome |
Ube3a |
Enhanced (↑p-mTOR, ↑p-S6K, ↑p-S6) |
Rapamycin
|
[110,117]
|
3.2.2. Neurofibromatosis type 1
Neurofibromatosis type 1 (NF1) is an autosomal dominant genetic condition characterized by the formation of neurofibromiomas and other nerve tumors [124]. In addition, NF1 patients also present cognitive impairments, as well as an increased susceptibility to suffer ASD [125,126]. NF1 is caused by mutations in the NF1 gene coding the protein neurofibromin, a Ras-GTPase activating protein leading to Ras signaling inhibition [127]. NF1 loss-of-function mutations lead to an enhanced Ras activity, increasing both PI3K-mTORC1 (Table 2) and Ras-Raf-MEK-ERK signaling (Figure 2). Interestingly, inhibitors of ERK have demonstrated to recover the neurological defects of NF1 mice [128,129]. In addition, lovastatin, a 3-hydroxy-3-methyl-glutaryl-coenzyme A reductase inhibitor, has been shown to ameliorate neurological deficits in this disorder through Ras inhibition [130,131]. Interestingly, a similar approach in the fragile X syndrome mouse model revealed a decrease in protein synthesis and reduced epileptogenesis [132]. These results suggest that the pharmacological reduction of Ras activity is a relevant therapeutic approach worth exploring in the context of mTORC1 signaling deregulation.
3.2.3. PTEN-related disorders
Loss of PTEN results in familial hamartoma-tumor syndromes and brain disorders [133] associated to autism-like conditions [134,135]. PTEN is a lipid dual-specificity phosphatase that converts PIP3 to PIP2 reducing the activity of PI3K-mTORC1 pathway [136]. In the absence of PTEN function, mTORC1 activity becomes significantly increased (Figure 2). The enhancement in protein translation due to mTORC1 hyperactivation (Table 2), leads to the autism phenotype of PTEN-related conditions [105]. Rapamycin was found useful in PTEN-deficient mice as it improved the autistic-like condition in these animals [115] pointing to a therapeutic relevance of mTORC1 blockade [212].
3.2.4. Fragile X syndrome
FXS patients are characterized by their intellectual disability trait. In some patients it is accompanied by hyperactivity, hypersensitivity to sensorial stimuli, attention deficits and autistic behavior [137]. FXS is caused by an accumulation of CGG repeats on the untranslated region of the FMR1 (fragile X mental retardation 1) gene that causes the silencing of FMRP (fragile X mental retardation protein) expression, a RNA-binding protein [58]. In normal conditions, FMRP binds many different mRNA molecules [138], so in its absence, there is a broad translational deregulation of the dendritic transcriptome [138,139]. Some of the mRNA molecules under FMRP regulation are involved in mTORC1 cascade, such as the p110β subunit of PI3K and PIKE, a PI3K enhancer [140]. Then, the loss of FMRP leads to the de-repression of p110β and PIKE mRNAs, which results in an increased PI3K-mTORC1 signaling [141,213]. Other FMRP mRNA targets participating in PI3K-mTORC1 pathway are Homer1a, PSD-95, eIF4A, eIF4G, NMDA receptor and mGlu (metabotropic glutamate) receptor [142,143]. Therefore, in FMRP scarcity, translation rates increase leading to the over-activation of mTORC1 signaling pathway [107,141] (Figure 2). The global result of FMRP absence is a higher basal protein synthesis due to the lack of translation repression, and the overstimulation of mTORC1 [107,144]. Interestingly, FMRP deficiency has been associated to deficits in activity-dependent synapse elimination due to ubiquitin-proteasome system alterations in the processing of PSD-95 [145]. Many approaches have been experimentally tested in FXS mouse models, as reviewed in ref.146. Among those, the mGlu5 receptor antagonist AFQ056 (mavoglurant) failed in phase Ⅱ clinical trial [147], indicating the need for additional research on other potential therapeutic approaches.
3.2.5. MECP2 disorders
There are two severe neurological disorders characterized by intellectual disability and autism: Rett syndrome [148,149] and MECP2 duplication syndrome [109,149]. On the one hand, Rett syndrome is caused by mutations in MECP2 (methyl-CpG-binding protein 2), an X-linked gene encoding a methylated DNA-binding protein that regulates gene expression and chromatin structure/function as a transcription activator and repressor [150]. Data from the Rett syndrome mouse model, the Mecp2 knockout mouse, mTORC1 pathway is down-regulated (Table 2), causing an abnormal synapse function [108]. On the other hand, MECP2 duplication is also responsible for a severe intellectual disability, ASD and developmental regression [109]. The mouse model for this disorder shows mTORC1 hyperactivity (Table 2), as well as an increase in spine turnover and dendritic growth [151]. Therefore, MeCP2 protein function results critical for synaptic function affecting mTORC1 activity.
3.2.6. Angelman syndrome
Algelman syndrome (AS) is characterized by severe developmental delay, language and cognitive deficits, unusual happy conduct, epilepsy and autistic like behavior [152]. AS is caused by the deficit in expression of the maternally inherited UBE3A gene [153]. In most tissues both copies of UBE3A are expressed. However, in neurons only the expression of the maternal copy is favored due to genomic imprinting [154]. The encoded protein, ubiquitin-protein ligase E3A, transfers ubiquitin from an E2 ubiquitin-conjugating enzyme to the target protein. Several target proteins have been described for ubiquitin-protein ligase E3A: ECT2 (epithelial cell transforming sequence 2 oncogene) [155], p53 [156], p27 [157], HR23A [158], Arc [159] and ephexin-5 [160]. Patients show a decrease in dendritic spine density [161] similar to what occurs in the animal model of the disorder, the UBE3A knockout mouse [162]. Interestingly, mTORC1 over-activation is observed in the animal model of the disorder (Table 2), and pharmacological inhibition with rapamycin manages to improve motor coordination and learning and memory in this mouse model [110,117].
4. Conclusions
The maintenance of protein homeostasis involves several complementary mechanisms of protein synthesis and degradation. Neurons have specific mechanisms to support proteostasis at synapses, and proteostasis deregulation has been pinpointed as a common factor involved in a wide range of central nervous system pathologies, including ASD. Different forms of ASD share common features that converge in alterations of the mTORC1 signaling pathway. Indeed, its over-activation, as well as its under-activation results in pathological consequences that converge at the synapse. The mTORC1 pathway plays a key role in synaptic proteostasis by regulating mRNA targeting and stability, translation initiation and progression, proteasome-mediated protein degradation and autophagy; therefore, the understanding of proteostasis regulation via mTORC1 and associated Ras signaling pathways are key to define the synaptic pathophysiology of ASD. However, the role played by mTORC2 is less well understood. This is due, in part, to the lack of specific inhibitors for mTORC2 that would allow assigning relative roles of both complexes in mTOR-dependent signaling. In addition, the interplay between these two complexes is not well established in the brain, especially under those pathological conditions where mTORC1 is constitutively over-activated, as in the cases summarized in the present review. The study of specific mTORC2 inhibitors (when available), dual inhibitors for mTORC1/mTORC2, as well as specific activators of these complexes, together with the use of genetic tools, will establish the foundations for the better understanding of this signaling pathway as a therapeutic target. The identification of the de-regulated features in mTORC1 pathway at synaptic sites and the fact that this signaling pathway can be pharmacologically targeted with specific inhibitors already available, open the possibility of addressing the synaptic alterations found in different disorders by targeting a single common pathway. This possibility that has been already explored in mouse models of TS, FXS and AS, may indeed be assessed in the clinical context in the near future, given the availability of the specific inhibitors of mTORC1 and the experience accumulated in their clinical use.
In this review, we have summarized the main studies that highlight the relevance of mTOR pathway in proteostasis and ASDs. Hence, the advances in the understanding of potentially common mechanisms at the proteostasis mechanisms are important to identify and develop novel powerful therapeutic approaches that may target shared affected mechanisms.
Acknowledgements
This review was supported by grant BFU2015-68568-P (MINECO/FEDER, EU) to AO.
Conflict of Interest
The authors declare no conflicts of interest in this paper.









 DownLoad:
DownLoad: