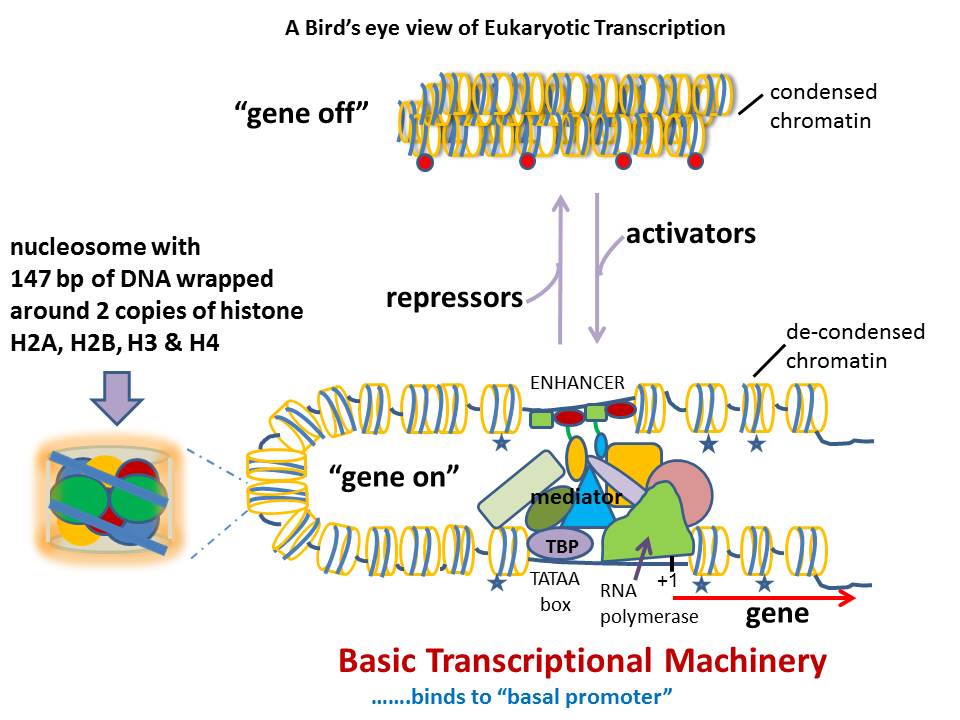
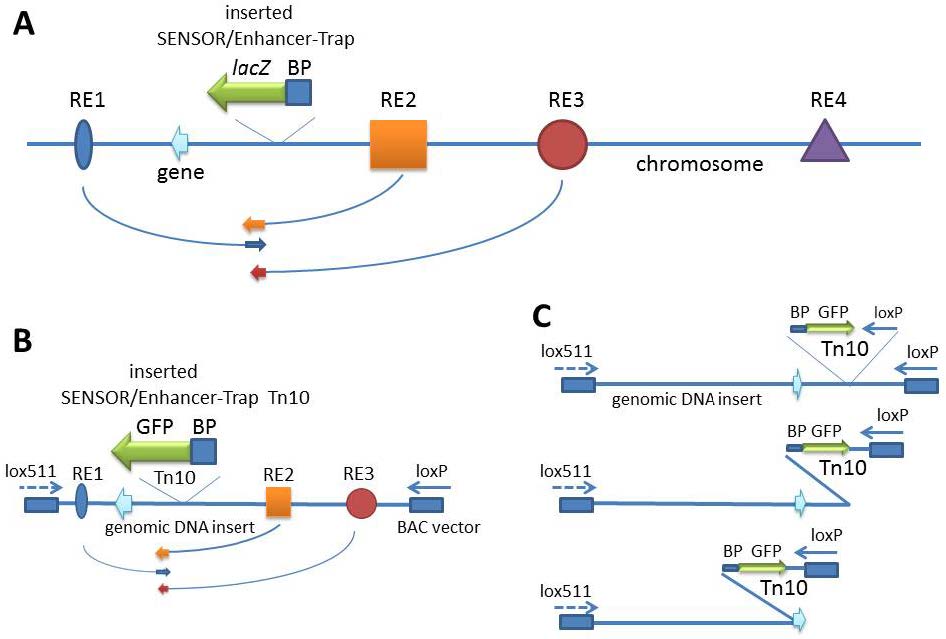
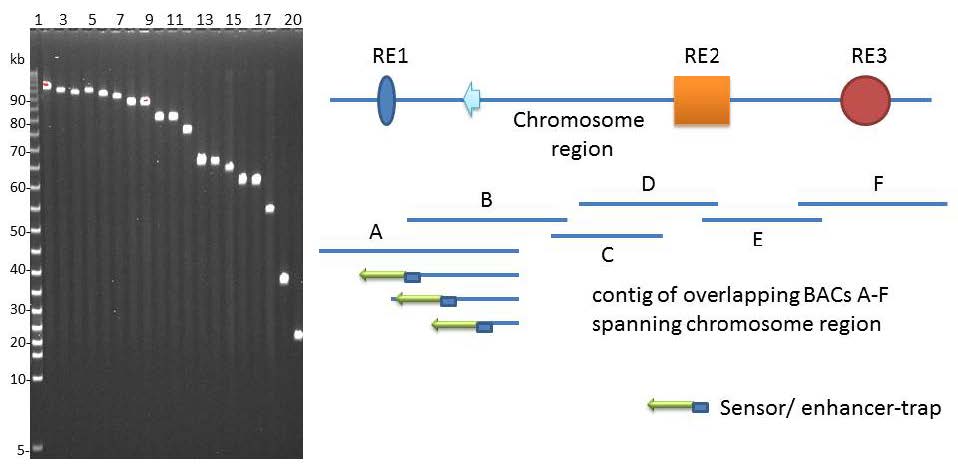
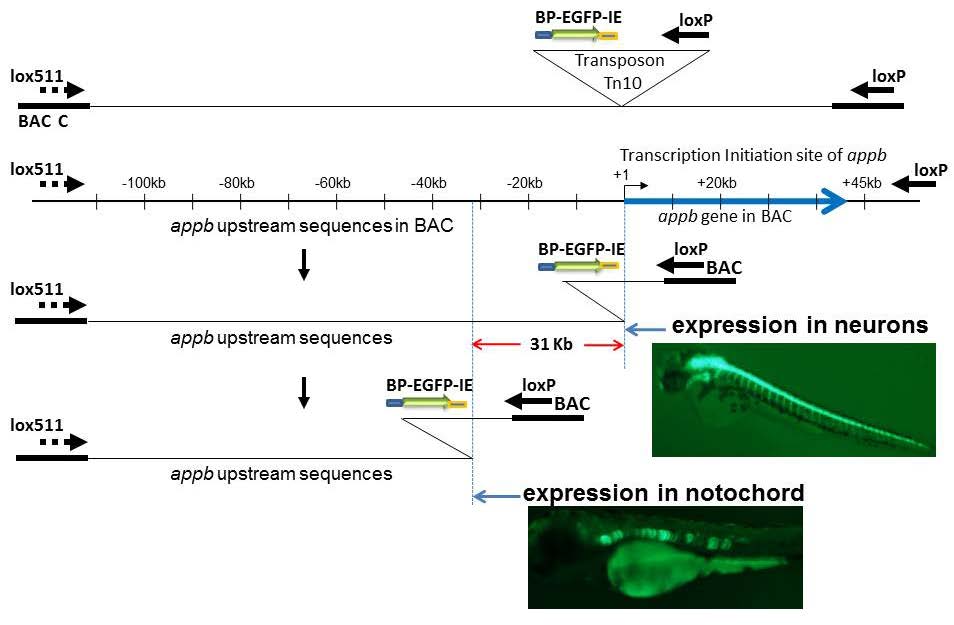
There is renewed interest in understanding expression of vertebrate genes in their chromosomal context because regulatory sequences that confer tissue-specific expression are often distributed over large distances along the DNA from the gene. One approach inserts a universal sensor/reporter-gene into the mouse or zebrafish genome to identify regulatory sequences in highly conserved non-coding DNA in the vicinity of the integrated reporter-gene. However detailed mechanisms of interaction of these regulatory elements among themselves and/or with the genes they influence remain elusive with the strategy. The inability to associate distant regulatory elements with the genes they regulate makes it difficult to examine the contribution of sequence changes in regulatory DNA to human disease. Such associations have been obtained in favorable circumstances by testing the regulatory potential of highly conserved non-coding DNA individually in small reporter-gene-containing plasmids. Alternative approaches use tiny fragments of chromosomes in Bacterial Artificial Chromosomes, BACs, where the gene of interest is tagged in vitro with a reporter/sensor gene and integrated into the germ-line of animals for expression. Mutational analysis of the BAC DNA identifies regulatory sequences. A recent approach inserts a sensor/reporter-gene into a BAC that is also truncated progressively from an end of genomic insert, and the end-deleted BAC carrying the sensor is then integrated into the genome of a developing animal for expression. The approach allows mechanisms of tissue-specific gene expression to be explored in much greater detail, although the chromosomal context of such mechanisms is limited to the length of the BAC. Here we discuss the relative strengths of the various approaches and explore how the integrated-sensor in the BACs method applied to a contig of BACs spanning a chromosomal region is likely to address mechanistic questions on interactions between gene and regulatory DNA in greater molecular detail.
1.
Introduction
Banana is the most cultivated fruit in South America, constituting a staple food for local populations and an important commercial product for farming families and countries that export it [1,2]. Brazil is the largest producer and consumer of bananas on the continent, making up a volume of 6.8 mi t, with 99% of production turned to the domestic market [2]. However, in the productivity ranking, the country occupies the 7th position, with an average of 15 t.ha−1, and a difference of 25 t.ha−1 from Ecuador's yield, which reaches 40.3 t.ha−1 [2]. Banana cultivation in Brazil is carried out mainly by family farmers but yields on small properties are generally low [3]. The low fertility of the soil in the producing regions associated with the absence of agricultural practices and the low economic power of the farmers directly imply the low productivity of banana farming [3,4].
Fertilization is one of the fundamental practices to achieve the productive potential of banana cultivars. However, depending on the characteristics of the production system, the need for fertilizers can represent more than 50% of the total cost of the activity in its first year [5,6]. On the other hand, the intensive use of fertilizers can contaminate the soil and cause eutrophication of water bodies [7,8]. Non-tanned poultry litter, for example, can cause the spread of pathogenic microorganisms and contamination of the soil with veterinary products residues [9,10]. The non-renewable character of synthetic fertilizers should be considered, whose sources are running out, such as phosphorus reserves [11]. Therefore, technological solutions that optimize the use of fertilizer by the plants can improve the productive efficiency of the banana crop and make it a more sustainable activity.
The use of plant biostimulants is a strategy that reduces the dependence on fertilizers as it optimizes the use of these inputs by crops. Biostimulants are products-based substances and/or microorganisms that increase the nutritional efficiency, tolerance to abiotic stress, and/or quality of crops, with dissociated effects on the nutritional content of products [12]. The genus Trichoderma has been widely studied for its multi-benefits, which include strains capable of acting as biocontrol agents and/or as biostimulants, with mechanisms that mainly result in the promotion of plant growth and/or reduction of the effects of abiotic stresses [13]. Biostimulation by Trichoderma involves synergistic mechanisms linked to the development of the root system, such as the production of rhizospheric compounds and the induction of phytohormones; the availability of nutrients, via nitrogen fixation, phosphorus, and potassium solubilization, mineralization of organic matter and secretion of enzymes and siderophores and; to plant defense, such as the secretion of antimicrobial substances and the induction of systemic tolerance and modulation of plant defense mechanisms [13,14,15].
In banana cultivation, Trichiderma promotes plants growth and reduces the incidence of Fusarium oxysporum F. sp. cubense [16]. In maize [17], mustard and tomato [18] and soybean [19], the inoculation of Trichoderma reduced the use of fertilizers by 50%, without harming the production, demonstrating the potential of the microorganism to optimize the use of fertilizers in different cultures. However, the effects of the use of microorganisms in suboptimal nutritional regimes are still unknown in the banana crop.
The economic dimension of technologies is an aspect increasingly explored in agricultural research, it is crucial to the decision of farmers to join or not to new practices. The economic viability diagnosis is a study that shows whether an activity will bring returns to the investor or not while comparing the costs of its implementation and operation with the revenues and benefits earned during a certain period [20]. Some works address the economic viability of different nutrient management in banana production [5,6,21,22]. Nonetheless, the use of Trichoderma-based biostimulant is a practice that has not been tested in the nutritional management of bananas yet, either in Amazonian field conditions. Moreover, for family farmers interested in investing in banana plantations, it is essential to be based on the costs and profitability of similar production systems to reduce the financial risks of the operation.
Knowing that Trichoderma promotes growth in plants and improves the availability and absorption of nutrients, the following questions were raised: (1) Using the Trichoderma-based biostimulants could reduce the fertilizer doses by 50% without harming the fruit production? (2) Could the uses of biostimulant improve the banana production performance under the 100% fertilizers doses? (3) Is the nutritional management of banana trees with the biostimulant economically viable in the family production system? Thereby, the objective of this study was to evaluate the agronomic performance and economic viability of the cultivation of bananas under fertilization practices with native Trichoderma in the family production system in the Amazon region.
2.
Materials and Methods
2.1. Characterization of the area
The study was conducted on the banana tree plantation in a family property, located in Ourém (1°33'02.8"S, 47°06'49.9"W), state of Pará, Brazil. The local climate fits on the Am type (hot and humid) of the Köppen classification, with a dry season between September and November. It has an annual rainfall of 2,400 mm, relative humidity of 83%, and a 26.2 ℃ annual average temperature.
The experimental area covers 355 m2, on the edge of a 6.3 ha banana tree plantation, where there was a secondary vegetation before. It has a flat relief, with a slight slope and a soil characterized as a yellow oxisol of sandy texture. The chemical and physical soil analysis presented the following results (0 to 20 cm layer): sand = 85.2%, silt = 4.1%, clay = 10.7%; pH in water = 5.56 (medium acidity), P (Mehlich) = 1.83 mg.dm−3 (very low), K+ = 0.07 cmolc.dm−3 (very low), Ca2+ = 2.05 cmolc.dm−3 (medium), Mg2+ = 0.35 cmolc.dm−3 (low), Al3+ = 0.21 cmolc.dm−3 (low), H + Al = 3.4 cmolc.dm−3 (medium), M.O. = 20.87 g.dm−3 (adequate), CTC (effective) = 2.68 cmolc.dm−3 (medium), CTC (potential) = 5.87 cmolc.dm−3 (medium), V = 42.08% (low), m = 7.84% (low – not harmful).
2.2. Implementation of the area and experimental design
The plantation occurred in December 2017. Banana tree seedlings Musa (AAAB group) 'BRS Pacoua', previously named PV 0376 were used. The seedlings (chifrinho type) were obtained in a commercial banana tree plantation, with plants less than 4 years old, which matrices presented excellent vigor and phytosanitary quality. The spacing of 4.5 m x 2.25 m x 2.25 m (1.035 plants.ha−1) was used, an arrangement used in association with açaí. At the time of planting, the pits were fertilized with 5 liters of poultry litter, 250 g of NPK (10-28-20), 100 g of potassium chloride (KCl), and 100 g of dolomitic limestone.
The experimental design was in randomized blocks, with 4 blocks, 4 replications per block, and 3 treatments, adding up to 48 experimental units. The following nutritional managements were tested: 100% F (control): 100% of fertilizers (synthetics and organics); 50% F + TA: 50% of fertilizers + T. asperellum (a mix of the isolates Ufra.T06, Ufra.T09, Ufra.T12, Ufra.T52) and; 100% F + TA: 100% of fertilizers + T. asperellum (a mix of the isolates).
The complete fertilization (100%F – control) referred to the usual fertilization of the partner farmer, being 15 liters of poultry litter, 900 g of NPK (10-28-20), and 300 g of KCl per clump, during the formation and; 15 liters of poultry litter and 900 g of NPK (10-28-20), per clump, in the production phase. The fertilizers doses were split into three times and applied during the year. The fertilization that was used is nearly equivalent to 83% of N, 220% of P2O5 and 102% of K2O recommended for the crop, considering the organic fertilization (available nutrients in the application year), and minerals performed at the planting, formation, and production [23].
2.3. Characterization, production, and inoculation of T. asperellum
The microorganisms that were used are part of the Plants Protection Laboratory (LPP) of the Federal Rural University of Amazonia, State of Pará, Brazil. Preparation based on four isolates of T. asperellum (Ufra.T06, Ufra.T09, Ufra.T12, Ufra.T52) was used, obtained from rhizospheric soils from reforested and natives Amazon forest areas [24]. The molecular identification of the strains was performed by de Sousa et al. [25].
The isolates of T. asperellum were grown separately in a PDA medium (potato, dextrose, and agar) and incubated for 96 h, under 28 ℃. From each isolate, were preparing an aqueous suspension at 108 conidia.mL−1. The suspensions were mixed and inoculated in autoclaved rice, incubated for 7 days, under 28 ℃. Starting from the colonized rice, the biostimulant was elaborated, an aqueous solution at 2 g.L−1 of autoclaved rice. In the field, each seedling received 300 mL of biostimulant, 100 mL at 30, 60, and 90 days after the planting. Additionally, 3 L of bioproduct were applied per clump.year−1, in a split way, 20 days after the fertilization operations.
2.4. Banana tree plantation management and production evaluation
The maintenance of the area consisted of the following operations: crowning, that is, cleaning around the clump; synthetic and organic fertilization application, in a semicircle (radius about 0.50 m), in front of the daughter plant; biostimulant application by irrigation, around the clump (radius about 0.50 m); thinning, with the maintenance of three plants per clump; defoliation, with the removal of senescent leaves; mowing, to control weed competition; cut off the banana inflorescence, under the last bunch, at a distance of 0.10 to 0.20 m and; irrigation, carried out from September to December, via conventional sprinkling, twice a week, in the amount of 30 liters per clump. Any phytosanitary control was performed.
The data collection occurred during the harvest of the 1st cycle of the culture, between November 2018 and December 2019. Harvests were carried out manually when the fruits reached the stage of maturity 2 (light green peel). The analyzed variables were: the numbers of hands per bunch (NHB) and the number of fingers per bunch (NFB), obtained from counting; the number of fingers per bunch, given by the difference between NFB and NHB; fruit length, the average length of 3 fruits of the 3rd bunch, measured with measuring tape; fruit diameter, the average diameter of 3 fruits from the 3rd bunch, measured with a caliper, in the central area of the fruits; the mass of the hands, the difference between the mass of the bunch (without central stalk) and NHB; bunch mass (with central stalk), with the cut of the peduncle 20 cm above the 1st bunch and; productivity (t.ha-1), based on the mass of the bunch (with central stalk) and density of 1,035 plants per hectare. Fertilization productivity was obtained by performing the division of the productivity by the total nitrogen, phosphorus, and potassium used in one hectare since the planting. The results were submitted to variance analysis (ANOVA) and the averages were compared by Turkey tests, p < 0.10, using the R programming language.
2.5. Production cost analysis
Production costs survey of the managements were performed according to the structure used by the Agricultural Economics Institute [26]. In this method, we obtain the Total Operating Cost (TOC), given by the addition of the Effective Operating Cost (EOC) plus the "Other costs" item (referring to the 5% rate on the EOC value).
The EOC estimate for the 1st planting year referred to the addition of the input expenses and services of the following items: A) Nutritional management; B) The Implantation of the banana tree plantation; C) Irrigation system implantation; D) Irrigation system and; E) Cultural treatments. From the 2nd planting year, EOC was compounded by the items: A) Nutritional management; D) Irrigation system; E) Cultural treatments; F) Harvest and post-harvest. In addition to the energy cost and irrigation service, item "D" was composed of expenses with maintenance and depreciation of the irrigation system, respectively 2.5% and 10% of the value of its implementation. The costs were budgeted for one hectare and in one year. Monetary values were converted from real (R$\$$) to American dollar (US$\$$), using the 4.0301 commercial exchange rate for purchase, 2019 annual average, made available by the Institute of Applied Economic Research – IPEA. Inputs quotation was made in 2019, in the commerce of the municipality of Castanhal-PA, where most inputs are purchased. The daily value (US$\$$ 8.26) was calculated based on the current basic salary, being US$\$$ 247.64. Energy expenses of the (D) Irrigation system item were estimated based on the water pump energy consumption (4.1 kWh), in the annual use of the system, being 384 h (12 h irrigation period) and in the kWh value of US$\$$ 0.12891.
2.6. Profitability of the activity analysis
The cash flow for the three nutritional practices was made considering the 6 years horizon. The 1st year consisted of the crop formation phase and, the subsequent years, constituted the production phase. For each cash flow period, there is an annual productive value (PROD.A), gross revenue, TOC, gross margin and accumulate gross margin. In the 2nd year, tested managements PROD.A was used, calculated by multiplying the obtained productivity and number two, the average value of harvested bunches in the year. From the 3rd year, estimated values were adopted, based on the productivity growth trend of BRS Pacoua (PV 0307), recorded by Leite et al. [27].
The gross revenue refers to the expected revenue with the activity, obtained by multiplying PROD.A (t.ha−1) and the average value of fruit ton – based on the selling price practiced by the producer, is US$\$$ 6.20 per 15 kg box. The gross margin refers to the resulting margin after the liquidation of TOC, obtained by subtracting the gross revenue and TOC of the season [28]. On the other hand, the accumulated gross margin is given by the addition between the gross margin of the season and the accumulated gross margin of the last season.
Additionally, economic indicators that consider all the cash flow periods were calculated, namely: break-even point (BEP), net present value (NPV), internal return rate (IRR), cost-benefit ratio, and simple payback. The interest rate applied to the calculation of the indicators was 3% per year, referring to the PRONAF Custeio credit line, directed to the family farm producers.
The break-even point (BEP) is the minimum of productivity to cover the TOC of the season, obtained by the difference between TOC and the average value of the fruit ton [28].
According to Samanez [29], the NPV refers to the present value of cash flows (benefits minus disbursements) generated by the activity throughout its life. The calculation is made from the expression:
In which I represents the initial investment; CF is the cash flow in the t period, being the results of subtraction gross revenue between TOC of a year; and K is the used discount rate, in this case, 3% per year. If the NPV > 0, activity is considered economically viable. For comparison purposes, the NPV results registered in other works went through a monetary correction for December 2019, based on the General Price Index - Internal Availability (IGP-DI), through the tool "Calculadora do Cidadão", made available by the Central Bank of Brazil. The monetary values were converted from real (R$\$$) to American dollar (US$\$$), using the commercial exchange rates for the purchase of 4.0301, the annual average of 2019, made available by IPEA.
The internal rate of return (IRR) is the investment returning rate [29]. Mathematically, IRR consists of the interest rate that annuls a NPV cash flow, satisfying the following equation:
As a decisive criterion, the enterprise is economically viable if IRR > K (interest rate used in the NPV calculation).
The cost-benefit ratio (CBR) that covers every cash flow year is obtained by the difference between the current income value (gross revenue) and the value of the current debt (TOC), both discounted at a given rate (3% per year). If this difference is superior to one, the activity is considered viable, it is less than one, and it is unviable because the revenues do not cover the cost of capital.
According to Bordeaux-Rego [30], the simple payback method refers to the invested capital returning time. In another way, consists of the period of cash flow in which the accumulated Gross Margin equals the value of the initial investment. Thereby, the lower the payback value, the greater the economic viability of the activity. In the decision process, the indicator is compared to the maximum term supported by the investor.
3.
Results and discussion
3.1. Agronomic performance of banana tree
Inserting T. asperellum in the nutritional management positively influenced the productivity parameters of the 1st cycle of the BRS Pacoua banana (Table 1). The combination of 100%F + TA provided an 11% increase in the numbers of hands per bunch, 17% in mass of the hands, and 23% in mass of bunch and productivity, about control (100%F). On the other hand, plants treated with 50%F + TA showed productive parameters like the control plants (100%F). Among TA inoculated plants, the use of 100% of fertilizers resulted in an average increase of 22% in the variables mass of the bunch and productivity. The high efficiency of nutrient use was verified on the 50%F + TA treatment, which for every kg of N resulted in almost 30 kg of banana production, and for every kg of P and K resulted in more than 13 kg of banana. From another angle, when was used 100%F (control) was necessary twice more N, P, and K to produce the same amount of banana obtained in 50%F + TA.
The average productivity of the 1st BRS Pacoua banana cycle was 7.32 t.ha−1, lower than productivity recorded in experiments conducted in the North and Northeast region of Brazil, an average of 10 t.ha−1, considering the density of 1,035 plants ha-1 and fertilization according to the soil analysis [27,31]. The 26% reduction of cultivar productive potential may be related to the sandy texture of the soil combined with the practices of fertilization and liming in the family production system, such as using only 20% of recommended limestone to raise the base saturation to 60%, localized application of the corrective (in the planting pits) and the use of 83% of indicated N doses to the culture.
In the present study, inoculation of T. asperellum increased banana yield (100%F + AT) and improved fertilizer use efficiency (50%F + AT). Several studies indicate that inoculation with the fungus enhances the growth and productivity of agricultural species such as maize [17], rice [32], soybean [33], and banana [16]. In addition to promoting plant growth, Trichoderma can also act as a biocontrol agent, a versatility relevant to its effectiveness in different environmental circumstances. In banana plants, T. asperellum (PZ6) increased plant growth, root activity, and seedling defensive capacity against Fusarium oxysporum f. sp. cubense [16]. In lowland rice, the T. asperellum isolates used in the present study increased plant growth by 12% and productivity by 70%, as well as reduced the severity of rice sheath blight by 26% [32]. Like the present study, Trichoderma reduced the need for fertilizers in maize [17], tomato and mustard [18], and soybean [19] crops. The effects of the combination of T. asperellum together with different doses of fertilizers on BRS Pacoua bananas are unprecedented findings, however, the benefits of nutrient management with different combinations of organic, inorganic, and microbial biofertilizers in banana cultivation have already been evidenced [34,35,36,37]. A study conducted with plant growth-promoting rhizobacteria (PGPR) showed that the use of only 33% N plus inoculation of biofertilizers (based on Azospirillum brasilense or Bacillus sphaericus) provides similar productivity to the use of 100% N in banana trees [34]. Likewise, in a study carried out with eight banana cultivars, the treatments of 100% NPK and 50% NPK plus PGPR-based biofertilizer showed no significant difference in vegetative growth rates [37].
The success obtained with the Trichoderma-based biofertilizer is due to the fungus' ability to assist in plant growth directly and indirectly, through several mechanisms of action, such as: nitrogen fixation; phosphorus and potassium solubilization; mineralization of organic matter; secretion of siderophores, enzymes and antimicrobial substances and; stimulation of phytohormone production. Thus, the synergistic activity of Trichoderma influences both the increase in the development of the root system of plants and the availability of nutrients in the soil, which culminate in increased productivity and adaptability of crops in the face of biotic and abiotic stresses [13,14,15]. In this context, the potential of Trichoderma strains from Amazonian forest soils to produce different organic acids, essential for converting phosphorus present in the soil into readily available forms for absorption (di or monobasic phosphate) is known [19]. At the same time, the high efficiency of T. asperellum (Ufra.T06, Ufra.T09, Ufra.T12, Ufra.T52) may still be related to the origin of the isolates, as they are native to the region where they were applied, which favors the adaptation of microorganisms to field conditions, a fact confirmed by López-Valenzuela et al. [17].
In this context, microbial technology is an important strategy to make banana production more sustainable, for improving the plant's productive performance, and to mitigate the fertilizer dependency. Besides that, the technique is efficient in field conditions and compatible with adopted practices in amazon family production systems. The insertion of T. asperellum in the banana tree nutritional management has high potential, as it is a low-cost technique, easy to use, non-polluting, and capable of reducing inorganic fertilizers dependency and their impacts on the environment. The successive inoculations with the microorganism may bring additional benefits to the agroecosystem, such as soil pathogens antagonisms and activation of the plant immune system.
3.2. Economic analysis
Independent of the period, the lowest total operating cost (TOC) occurred in the 50%F + TA management, followed by 100%F (control) and 100%F + TA, a result motivated by the NPK proportions of the treatments and the use of biofertilizer (Table 2). In the year of implementation, the TOC of 50%F + TA was US$\$$ 4,781.46 per ha (US$\$$ 4.62 per clump) and, from the 2nd year, it was US$\$$ 1,733.17 per ha (US$\$$ 1.67 per clump). This treatment generated annual savings of US$\$$ 371.03, which represents 7.2% and 17.6% of the cost to supply 100% of the fertilizers in the 1st year and from the 2nd year, respectively. On the other hand, 100%F + TA resulted in a US$\$$ 190.05 annual increase in managerial TOC for both periods. In an irrigated system, Rodrigues et al. [5] calculated a TOC of US$\$$ 3,042.45 for the year of implementation and US$\$$ 2,093.62 for subsequent years (corrected values). In non-irrigated plantations, the costs are usually lower, even concerning the 50%F + TA treatment. Lacerda et al. [21] indicate a TOC of US$\$$ 1,271.03 in the year of implementation and US$\$$ 785.11 in the years of production (corrected values).
According to the percentage share of production costs of the TOC (Table 3), in the 1st year of cultivation, the expenses with the implantation of the banana plantation and the irrigation system were the most representative, regardless of the treatment adopted. From the 2nd year, most of the disbursements were spent on inputs and services related to nutritional management and the operationalization of the irrigation system (including the depreciation of equipment). Furthermore, the share of expenditure on nutritional management was lower in the area under 50%F + TA, 5.63 and 8.55 percentage points less than 100%F (control) in the 1st year and from the 2nd year onwards, respectively. On the other hand, with the control treatment, 100%F + TA generated an increase of 2.59 percentage points in the 1st year and 3.31 percentage points from the 2nd year onwards.
In a non-irrigated 'Prata-Anã' banana crop in southeast Goiás [5], costs with inputs represented 74.06% of the total cost (TOC) of the activity. However, the authors did not specify the types of inputs used. Campos et al. [6] evaluated the economic performance of fertilization management in 'Prata-Anã' Gorutuba banana, in the central region of Minas Gerais, and found that inputs correspond to the highest costs in the year of implementation (58.19% of the EOC), followed by costs with cultural and phytosanitary treatments (16.24%), soil preparation and planting (10.63%) and irrigation (9.21%), contradicting the trend indicated in the present study. Such differences are linked to some aspects of the production system described by Campos et al. [6], such as planting density (1,666 plants/ha), the types of fertilizers used, and the values attributed to the costs of implementing and maintaining the irrigation system.
The association between T. asperellum plus 100% or 50% of the fertilizers provided higher returns, according to the analysis of the NPV, IRR, B/C, payback, and BEP indicators (Table 4). The use of T. asperellum as a supplement to 100% fertilization increased the NPV by 28.8% compared to the control treatment, equivalent to a revenue of US$\$$ 8,944.40. At 50%F + TA, this gain was US$\$$ 1,936.35, 6.2% over standard management. Regardless of the management adopted, the initial investment is paid in the 3rd year of planting. However, after discounting the initial investment, the gross margin accrued up to the 3rd year is US$\$$ 8,548.32 for 100%F + TA and US$\$$ 6,877.74 for 50%F + TA, margins that are equivalent to the respective increment of 47.2% and 18.4% on the 100%F return (Table 4). These results are corroborated by the internal rate of return (IRR), or rate of return on invested capital, with the highest value of the IRR for 100%F + TA (133.56%), followed by the management 50%F + TA (126.15%) and by the control treatment (112.27%). Lacerda et al. [21] and Rodrigues et al. [5] corroborate that the banana crop has high IRR values (194% and 199%, respectively). These rates are higher than the cost of capital of the main lines of credit that finance family farming activities in the region, such as the Constitutional Fund for Financing in the North (FNO) and the National Program for Strengthening Family Agriculture (PRONAF), a fact that attests to the high profitability of banana farming.
The B/C ratio was higher than 1.00 for all treatments (Table 4), evidencing the economic viability of banana cultivation regardless of the practice adopted. However, the 50%F + TA management presented a greater share of revenues concerning the TOC, followed by the 100%F + TA management. In practice, for every dollar invested, you get US$\$$ 3.60 for 50%F + TA and US$\$$ 3.52 for 100%F + TA, benefits superior to the standard treatment margin, US$\$$ 3.10. On the other hand, the lowest leveling point (BEP), the minimum production necessary to cover the production cost, occurred in the management 50%F + TA (4.19 t.ha−1), followed by 100%F (5.09 t.ha−1) and 100%F + TA (5.55 t.ha−1), corroborating the results related to TOC. Thus, the profitability obtained with 50%F + TA is strongly related to the reduction of production costs, and the profitability of 100%F + TA to the increase in gross revenue, due to the increase in productivity.
In a 'Prata-Anã' banana plantation in northeastern Brazil, under conventional soil management and different irrigation depths, Barbosa et al. [22] recorded averages of 2.13 for B/C ratio, 147% for IRR, and US$\$$ 34,047.97 for NPV (corrected value). The average performance of the B/C ratio of the present study (3.41) was superior to that reported by the authors [22], a difference explained by the operating costs of the production systems compared, possibly higher in the work by Barbosa et al. [22], for the daily use of the irrigation system; as well as the estimated yields and marketing prices of bananas, US$\$$ 0.41 kg−1 in the present study and US$\$$ 0.27 kg−1 (corrected value) in the authors' work [22]. The average IRR of the present study (123.99%) was lower than that reported by the authors, possibly influenced by higher estimated yields and lower interest rate (2% per year) used in the calculation of economic indicators for 'Prata-Anã' banana. On the other hand, the average NVP of both works was similar.
The economic indicators demonstrated by Lacerda et al. [21], considering a 5-year cash flow, were NPV of US$\$$ 6,504.29 (corrected value), B/C ratio of 7.10, BEP of 2.58 t.ha−1, and capital recovery period of 2 years. In turn, Rodrigues et al. [5] recorded NPV for 5 years of US$\$$ 21,280.86 (corrected value), B/C ratio of 3.12, and payback in the 2nd year. Thus, the average NPV of the present study (US$\$$ 34,696.49) is about 5.3 and 1.6 times higher than the NPV obtained by Lacerda et al. [21] and by Rodrigues et al. [5], respectively. However, the results of the B/C, BEP, and payback ratio for Lacerda et al. [21] and, from payback to Rodrigues et al. [5], performed better than in the present study.
Such divergences are attributed to the costs inherent to the different production systems evaluated, the yields and interest rates considered, as well as to the time of cash flow and marketing prices.
In the present study, the use of T. asperellum combined with fertilization provided greater profitability for the family producer, either through the reduction of input costs (50%F + TA) or through the increase in revenues, due to the increase in productivity (100%F + TA). Thus, microbial technology proved to be an economically viable strategy, with high profitability, a fundamental aspect of the sustainability of the activity. Additionally, recorded economic indicators can serve as a basis for decision-making on planting planning, adoption of technologies, and contracting rural credit financing – a significant contribution, since the low capital contribution to invest in inputs remains one of the main obstacles to increasing banana productivity in the context of family farming.
4.
Conclusions
This study is the first to demonstrate that T. asperellum inoculation increases the productivity of banana trees fertilized with 100% fertilizers, as well as reducing by 50% the need for these inputs, keeping the plant's productivity. The microbial technology is economically viable, increasing the profitability of banana tree cultivation by reducing the expenses or incrementing the revenues, showing itself as an efficient strategy to improve the banana tree agronomic performance, increase the efficiency of fertilizer use, and raise the profits to the family farmer, especially in Amazon.
Acknowledgments
This work was carried out with the support of the Coordination for the Improvement of Higher Education Personnel in Brazil (CAPES), the Graduate Program in Agronomy-UFRA (PGAGRO), and the National Council for Scientific and Technological Development (CNPq). The authors thank colleagues, Rubens Leite, and Matheus Konno, for their help in revising the text, and the owners of the banana plantation where the research was carried out, for having provided the orchard for the experiment.
Conflict of interest
The authors declare no conflict of interest in this paper.









 DownLoad:
DownLoad: