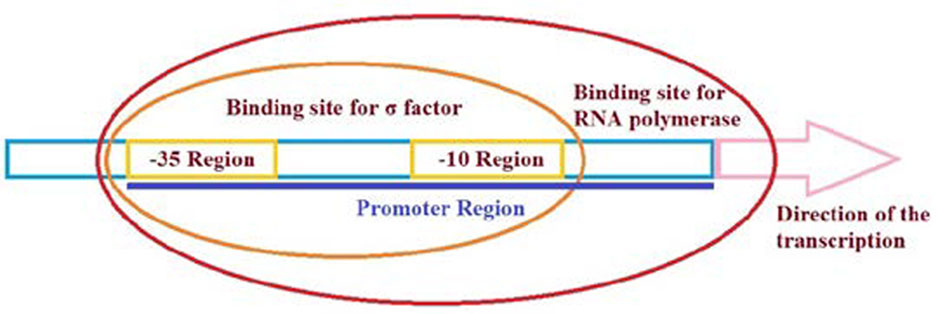
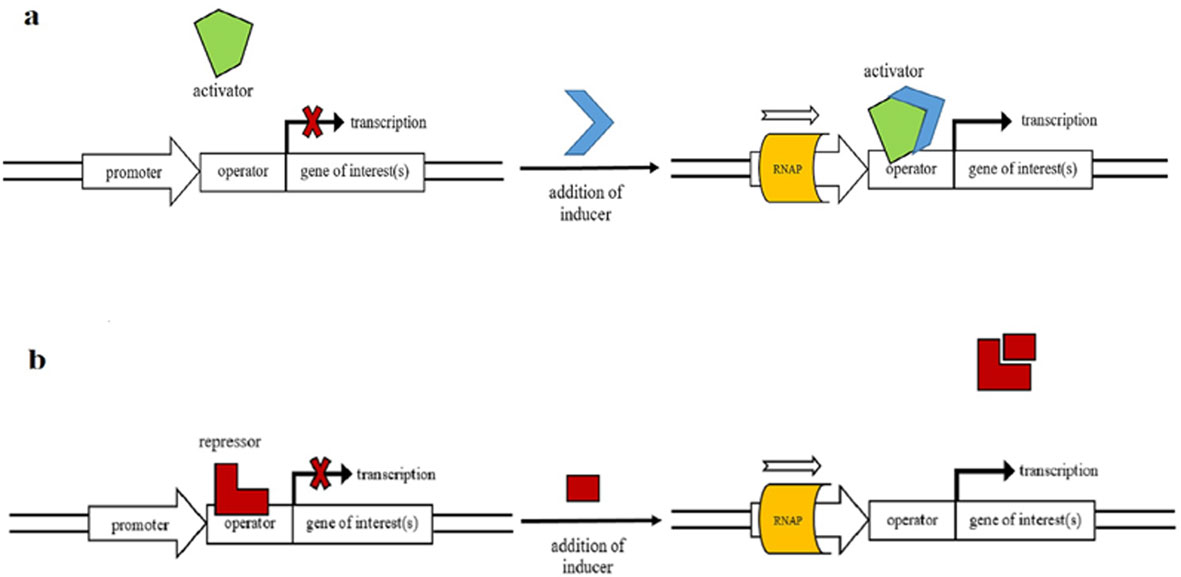
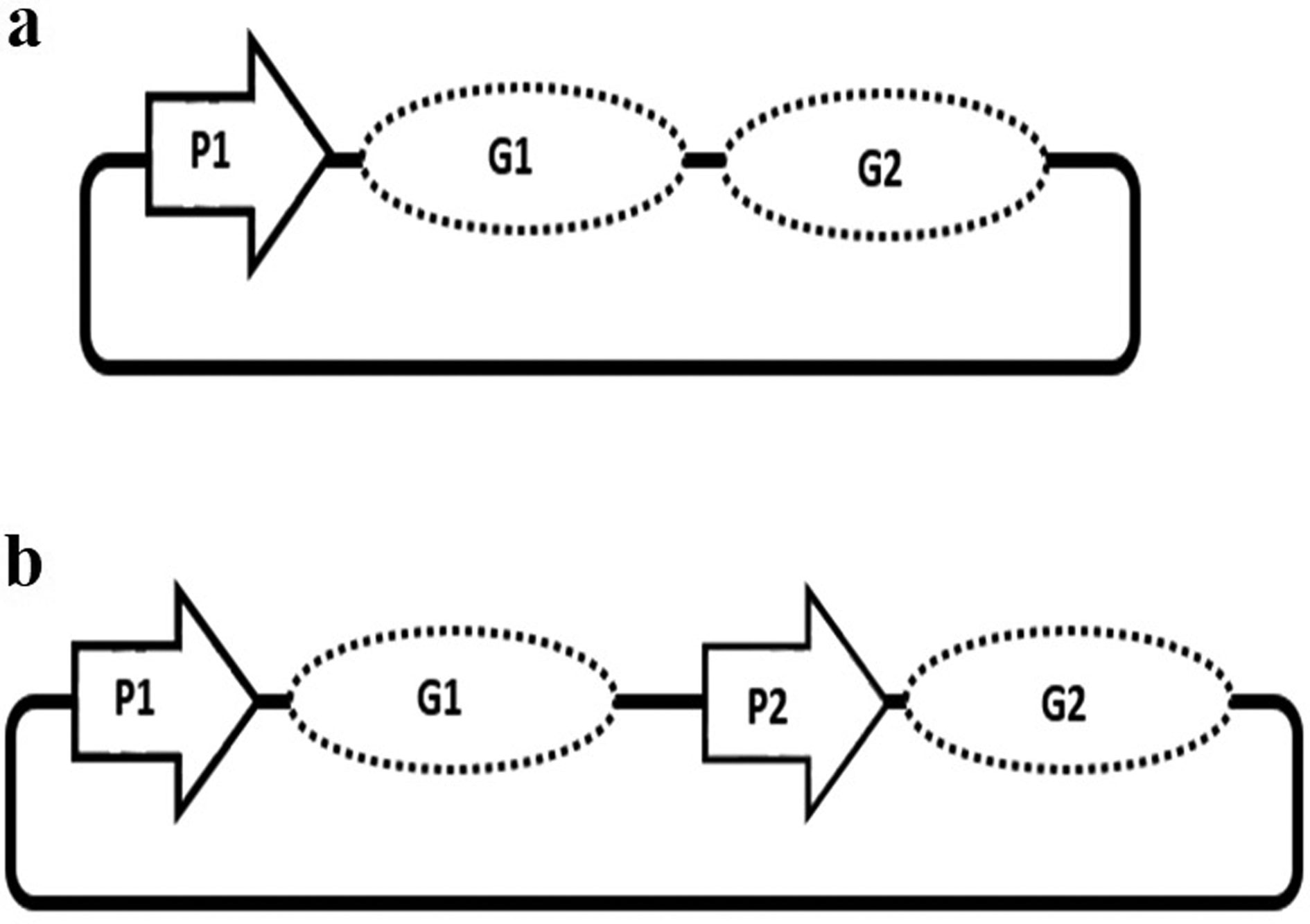
Recombinant proteins have an economical value with their utilization in many areas from food industry to pharmaceutical and chemical industry. Therefore, it is of great importance to establish optimum production systems for the proteins of interest. One of the critical steps in protein production is regulation of the gene expression. Promoters are among the key regulatory elements which can directly control the level of recombinant gene expression in a host cell. Thus, a suitable promoter is required for optimum gene expression. Promoter engineering is an innovative approach to find out the best promoter system for the expression of recombinant genes, which influences the overproduction of proteins of interest. In this review, some of the bacterial hosts highly used in recombinant protein production were discussed. Next, the importance of promoters in recombinant gene expression, and promoter engineering for enhanced protein production were described. Utilization of double promoter systems was highlighted as one of the successful techniques in overproduction of recombinant proteins. Increment in the variety and availability of the novel methodologies especially in the synthetic biology is expected to increase the quality and the quantity of recombinant proteins with an economical value.
1.
Introduction
In [1,2], Carlitz initiated study of the degenerate Bernoulli and Euler polynomials and obtained some arithmetic and combinatorial results on them. In recent years, many mathematicians have drawn their attention to various degenerate versions of some old and new polynomials and numbers, namely some degenerate versions of Bernoulli numbers and polynomials of the second kind, Changhee numbers of the second kind, Daehee numbers of the second kind, Bernstein polynomials, central Bell numbers and polynomials, central factorial numbers of the second kind, Cauchy numbers, Eulerian numbers and polynomials, Fubini polynomials, Stirling numbers of the first kind, Stirling polynomials of the second kind, central complete Bell polynomials, Bell numbers and polynomials, type 2 Bernoulli numbers and polynomials, type 2 Bernoulli polynomials of the second kind, poly-Bernoulli numbers and polynomials, poly-Cauchy polynomials, and of Frobenius-Euler polynomials, to name a few [3,14,16,17,18] and the references therein. They have studied those polynomials and numbers with their interest not only in combinatorial and arithmetic properties but also in differential equations and certain symmetric identities [4,5] and references therein, and found many interesting results related to them [12,19,20,21,22,23,24,25,26,27,28]. It is remarkable that studying degenerate versions is not only limited to polynomials but also extended to transcendental functions.
The Bernoulli polynomials of the second are defined by as follows (see [9,13])
When x=0, bq(0)=bq are called the Bernoulli numbers of the second kind.
The degenerate exponential function exλ(z) is defined by (see [6,7,8,9,10,11,12,13,14,15,16,17,18,19])
We note that
where (x)q,λ=x(x−λ)⋯(x−(q−1)λ),(q≥1), (x)0,λ=1.
Note that
The degenerate Bernoulli polynomials which are defined by Carlitz's as follows (see [1,2])
At the point x=0, βq(λ)=βq(0;λ) are called the degenerate Bernoulli numbers.
Note that
The polylogarithm function is defined by
Note that
The poly-Bernoulli polynomials of the second are defined by (see [13])
In the case when x=0, b(k)q=b(k)q(0) are called the poly-Bernoulli numbers of the second kind.
The modified degenerate polyexponential function is defined by (see [14])
It is noteworthy to mention that
The degenerate poly-Genocchi polynomials which are defined by Kim et al. as follows (see [14])
When x=0, G(k)q,λ=G(k)q,λ(0) are called the degenerate poly-Genocchi numbers.
For λ∈R, Kim-Kim defined the degenerate version of the logarithm function, denoted by logλ(1+t) as follows (see [11])
being the inverse of the degenerate version of the exponential function eλ(z) as has been shown below
It is noteworthy to mention that
The degenerate Daehee polynomials are defined by (see [15])
In the case when x=0, Dq,λ=Dq,λ(0) denotes the degenerate Daehee numbers.
The degenerate Bernoulli polynomials of the second kind which are defined by Kim et al. as follows (see [9])
When x=0, bq,λ=bq,λ(0) are called the degenerate Bernoulli numbers of the second kind.
Note here that limλ→0bq,λ(x)=bq(x),(q≥0).
The degenerate Stirling numbers of the first kind are defined by
It is noticed that
are the Stirling numbers of the first kind presented by
The degenerate Stirling numbers of the second kind are defined by (see [8])
It is clear that
are the Stirling numbers of the second kind specified by
Motivated by the works of Kim et al. [11,14], in this paper, we study the type 2 degenerate poly-Bernoulli polynomials of the second kind arising from modified degenerate polyexponential function and obtain some related identities and explicit expressions. Also, we establish the type 2 degenerate unipoly-Bernoulli polynomials of the second kind attached to an arithmetic function by using modified degenerate polyexponential function and discuss some properties of them.
2.
Type 2 degenerate poly-Bernoulli polynomials of the second kind
Here, the type 2 degenerate poly-Bernoulli polynomials of the second kind are defined by using the modified degenerate polyexponential function which is called the degenerate poly-Bernoulli polynomials of the second kind as
When x=0, Pb(k)j,λ=Pb(k)j,λ(0) are called the type 2 degenerate poly-Bernoulli numbers of the second kind.
Note that
where Pb(k)j(x) are called the type 2 poly-Bernoulli polynomials of the second kind (see [9]).
First, we note that
By making use of (2.1) and (2.3), we see that
Therefore, by (2.3) and (2.4), we obtain the following theorem.
Theorem 2.1. For k∈Z and j≥0, we have
Corollary 2.1. Putting k=1 in Theorem 2.1 yields
Let 1≤k∈Z. For s∈C, the function χk,λ(s) is given as
From Eq (2.5), we have
For any s∈C, the second integral is absolutely convergent and thus, the second term on the r.h.s. vanishes at non-positive integers. That is,
On the other hand, the first integral in Eq (2.7), for ℜ(s)>0 can be written as
which defines an entire function of s. Thus, we may include that χk,λ(s) can be continued to an entire function of s.
Further, from (2.6) and (2.7), we obtain
In view of (2.8), we obtain the following theorem.
Theorem 2.2. Let k≥1 and m∈N⋃{0}, s∈C, we have
Using (1.8), we observe that
Thus, by (2.9), for k≥2, we get
From (2.1) and (2.10), we get
Therefore, by (2.12), we obtain the following theorem.
Theorem 2.3. For j∈N and k∈Z, we have
Corollary 2.2. Taking k=2 in Theorem 2.3 yields
Replacing z by eλ(z)−1 in (2.1), we get
On the other hand,
In view of (2.13) and (2.14), we get the following theorem.
Theorem 2.4. For k∈Z and j≥0, we have
By using (2.1), we get
Therefore, by comparing the coefficients on both sides of (2.15), we obtain the following theorem.
Theorem 2.5. For j≥0, we have
By making use of (1.3) and (2.1), we have
Therefore, by Eq (2.16), we obtain the following theorem.
Theorem 2.6. For j≥0, we have
By using (2.1), we have
On the other hand,
Thus, by equations (2.17) and (2.18), we get the following theorem.
Theorem 2.7. For j≥0, we have
From (2.1), we have
Therefore, by comparing the coefficients on both sides of (2.19), we obtain the following theorem.
Theorem 2.8. For j≥0, we have
3.
The degenerate unipoly-Bernoulli polynomials of the second kind
Let p be any arithmetic real or complex valued function defined on N. Kim-Kim [7] presented the unipoly function attached to polynomials p(x) as
Moreover,
represent the known ordinary polylogarithm function.
Dolgy and Khan [3] introduced the degenerate unipoly function attached to polynomials p(x) are considered as follows
We see that
is the modified degenerate polyexponential function.
Now, we introduce the degenerate unipoly-Bernoulli polynomials of the second kind attached to polynomials p(x) as
When x=0, Pb(k)j,λ,p=Pb(k)j,λ,p(0) are called the degenerate unipoly-Bernoulli numbers of the second kind attached to p.
If we take p(j)=1Γ(j). Then, we have
For k=1, we have
Thus, we have
By making use of (1.12) and (3.3), we note that
Thus, we have the required result.
Lemma 3.1. For k∈Z, we have
Recalling from (3.5), we have
Therefore, by comparing the coefficients on both sides of (3.9), we obtain the following theorem.
Theorem 3.1. For j≥0 and k∈Z. Then
Moreover,
Using (3.5), we have
Upon comparing the coefficients on both sides of Eq (3.10), we get the following theorem.
Theorem 3.2. For j≥0 and k∈Z. Then
By making use of (1.11), (1.12) and (3.5), we have
Thus, by comparing the coefficients on both sides of (3.11), we obtain the following theorem.
Theorem 3.3. For j≥0 and k∈Z. Then
4.
Numerical computations
In this section, certain numerical computations are done to calculate certain zeros of the degenerate poly-Bernoulli polynomials of the second kind and show some graphical representations. The first five members of Pb(k)j,λ(x) are calculated and given as:
To show the behavior of Pb(k)j,λ(x), we display the graph Pb(k)j,λ(x) for k=4 and λ=3, this graph is presented in Figure 1.
Next, the approximate solutions of Pb(k)j,λ(x)=0 when k=4 and λ=3, are calculated and listed in Table 1.
The zeros of Pb(k)j,λ(x) for λ∈C,j=12 are plotted in Figure 2.
The stacking structure of approximate zeros of Pb(k)j,λ(x)=0 for λ=4,j=1,2,...,12 is given in Figure 3.
5.
Conclusions
In this article, we introduced the type 2 degenerate poly-Bernoulli polynomials of the second kind and derived many related interesting properties. Furthermore, we defined the degenerate unipoly Bernoulli polynomials of the second kind and established some considerable results. Finally, certain related beautiful zeros and graphs are shown.
Acknowledgments
The authors would like to express the gratitude to Deanship of Scientific Research at King Khalid University, Saudi Arabia for providing funding research group under the research grant number R G P.1/162/42.
Conflict of interest
The authors declare no conflict of interest.










 DownLoad:
DownLoad: